| General information about the entry |
| View entry in simple text format |
| Entry name | MDHC_RAT |
| Primary accession number | O88989 |
| integrated into World-2DPAGE Repository (0022) on | May 7, 2010 (release 1) |
| 2D Annotations were last modified on | June 22, 2011 (version 2) |
| General Annotations were last modified on | November 24, 2011 (version 2) |
| Name and origin of the protein |
| Description | RecName: Full=Malate dehydrogenase, cytoplasmic; EC=1.1.1.37; AltName: Full=Cytosolic malate dehydrogenase;. |
| Gene name | Name=Mdh1
Synonyms=Mdh
|
| Annotated species | Rattus [TaxID: 10114] |
| Taxonomy | Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; Eutheria; Euarchontoglires; Glires; Rodentia; Sciurognathi; Muroidea; Muridae; Murinae. |
| References |
| [1] |
2D-PAGE GEL CHARACTERIZATION
DOI=10.1002/pmic.201000593;
Burniston J.G., Kenyani J., Wastling J.M., Burant C.F., Qi N.R., Koch L.G., Britton S.L.
''Proteomic analysis reveals perturbed energy metabolism and elevated oxidative stress in hearts of rats with inborn low aerobic capacity''
Proteomics 11(16):3369-3379 (2011)
|
|
| 2D PAGE maps for identified proteins
|
|
How to interpret a protein
|
HCR_LCR_HEART {HCR LCR Cardiac Map}
Rattus
Tissue: Heart
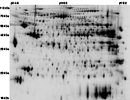
map experimental info
|
|
HCR_LCR_HEART
MAP LOCATIONS:
IDENTIFICATION: SPOT 348: SeqCov=37.7%. Peptides MS=12/20. Mascot PMF Score=103 [1]
SPOT 440: SeqCov=27.2%. Peptides MS=9/20. Mascot PMF Score=63 [1]; SPOT 513: SeqCov=32%. Peptides MS=10/20. Mascot PMF Score=77 [1]; SPOT 514: SeqCov=40.1%. Peptides MS=14/20. Mascot PMF Score=122 [1].
PEPTIDE EVIDENCE: SPOT 348: 221-GEFITTVQQR-230 (68) / 299-FVEGLPINDFSR-310 (77) / 180-NVIIWGNHSSTQYPDVNHAK-199 (95) [1]
SPOT 440: 221-GEFITTVQQR-230 (62) / 299-FVEGLPINDFSR-310 (88) [1]; SPOT 513: 221-GEFITTVQQR-230 (49) / 299-FVEGLPINDFSR-310 (69) [1].
MAPPING (identification):
|
| Copyright |
| Data from Dr. Jatin Burniston, Liverpool John Moores University, UK |
| Cross-references |
| UniProtKB/Swiss-Prot | O88989; MDHC_RAT. |