|
|
|
|
|
| Searching in 'World-2DPAGE Repository [0022]' for entry matching: P48721 |
World-2DPAGE Repository (0022): P48721
P48721:
Nice View - a user-friendly view of this entry
ID GRP75_RAT;
AC P48721;
DT 07-May-2010, integrated into [0022] - HCR LCR Cardiac Mapy (release 1).
DT 22-Jun-2011, 2D annotation version 2.
DT 24-Nov-2011, general annotation version 2.
DE RecName: Full=Stress-70 protein, mitochondrial; AltName: Full=75 kDa
DE glucose-regulated protein; Short=GRP-75; AltName: Full=Heat shock 70 kDa
DE protein 9; AltName: Full=Mortalin; AltName: Full=Peptide-binding protein
DE 74; Short=PBP74; AltName: Full=mtHSP70; Flags: Precursor;.
GN Name=Hspa9; Synonyms=Grp75, Hspa9a;
OS Rattus.
OC Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
OC Mammalia; Eutheria; Euarchontoglires; Glires; Rodentia; Sciurognathi;
OC Muroidea; Muridae; Murinae.
OX NCBI_TaxID=10114;
IM HCR_LCR_HEART.
RN [1]
RP 2D-PAGE GEL CHARACTERIZATION.
RX DOI=10.1002/pmic.201000593;
RA Burniston J.G., Kenyani J., Wastling J.M., Burant C.F., Qi N.R., Koch
RA L.G., Britton S.L.;
RT ''Proteomic analysis reveals perturbed energy metabolism and elevated
RT oxidative stress in hearts of rats with inborn low aerobic capacity'';
RL Proteomics 11(16):3369-3379 (2011).
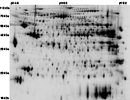
2D -!- MASTER: HCR_LCR_HEART;
2D -!- PI/MW: SPOT 785=5.97/74097;
2D -!- PI/MW: SPOT 790=5.97/74097;
2D -!- IDENTIFICATION: SPOT 785: SeqCov=20%. Peptides MS=12/20. Mascot PMF
2D Score=84 [1]; SPOT 790: SeqCov=11.9%. Peptides MS=7/20. Mascot PMF
2D Score=33 [1].
2D -!- PEPTIDE EVIDENCE: SPOT 785: 378-SDIGEVILVGGMTR-391 (104) /
2D 378-SDIGEVILVGGMTR-391 Oxidation (M) (48) / 499-LLGQFTLIGIPPAPR-513 (70)
2D [1]; SPOT 790: 378-SDIGEVILVGGMTR-391 (81) / 378-SDIGEVILVGGMTR-391
2D Oxidation (M) (37) / 499-LLGQFTLIGIPPAPR-513 (57) [1].
2D -!- MAPPING: SPOT 785: Peptide mass fingerprinting [1]; SPOT 785:
2D Tandem mass spectrometry [1]; SPOT 790: Peptide mass fingerprinting [1];
2D SPOT 790: Tandem mass spectrometry [1].
CC ---------------------------------------------------------------------------
CC Data from Dr. Jatin Burniston, Liverpool John Moores University, UK
CC ---------------------------------------------------------------------------
DR UniProtKB/Swiss-Prot; P48721; GRP75_RAT.
//
The following 2-D map is available for this protein:
|
HCR LCR Cardiac Map
 |
|---|
World-2DPAGE Repository (search AC)
|