Attention: World-2DPAGE is no longer maintained.
World-2DPAGE Repository no longer accepts submissions.
[Click to access protein entry]
Spot: 654 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 654 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 524 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 41523
================
*MK14_RAT*
accession n°: MK14_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 524 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 41523
================
*MK14_RAT*
accession n°: MK14_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 413 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 413 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 1000 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 53757
================
*VIME_RAT*
accession n°: VIME_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 1000 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 53757
================
*VIME_RAT*
accession n°: VIME_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 575 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 575 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 184 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 38757
================
*FETUA_RAT*
accession n°: FETUA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 184 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 38757
================
*FETUA_RAT*
accession n°: FETUA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 913 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 913 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 106 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 106 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 170 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 74097
================
*GRP75_RAT*
accession n°: GRP75_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 170 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 74097
================
*GRP75_RAT*
accession n°: GRP75_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 260 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 53757
================
*VIME_RAT*
accession n°: VIME_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 260 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 53757
================
*VIME_RAT*
accession n°: VIME_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 706 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 29326
================
*1433E_RAT*
accession n°: 1433E_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 706 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 29326
================
*1433E_RAT*
accession n°: 1433E_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 314 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 314 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 212 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 212 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 60 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 83571
================
*HS90B_RAT*
accession n°: HS90B_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 60 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 83571
================
*HS90B_RAT*
accession n°: HS90B_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 573 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 573 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 835 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 30100
================
*APOA1_RAT*
accession n°: APOA1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 835 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 30100
================
*APOA1_RAT*
accession n°: APOA1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 51 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 51 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 499 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 44429
================
*APOA4_RAT*
accession n°: APOA4_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 499 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 44429
================
*APOA4_RAT*
accession n°: APOA4_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 726 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 28456
================
*1433G_RAT*
accession n°: 1433G_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 726 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 28456
================
*1433G_RAT*
accession n°: 1433G_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 427 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 427 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 782 (rattus_soleus-dige_4.5-9.5)
pI: 9.30 Mw: 25360
================
*GSTA3_RAT*
accession n°: GSTA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 782 (rattus_soleus-dige_4.5-9.5)
pI: 9.30 Mw: 25360
================
*GSTA3_RAT*
accession n°: GSTA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 534 (rattus_soleus-dige_4.5-9.5)
pI: 6.50 Mw: 40044
================
*IDH3A_RAT*
accession n°: IDH3A_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 534 (rattus_soleus-dige_4.5-9.5)
pI: 6.50 Mw: 40044
================
*IDH3A_RAT*
accession n°: IDH3A_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 837 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 837 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 870 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 870 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 679 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 679 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 116 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 116 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 661 (rattus_soleus-dige_4.5-9.5)
pI: 10.00 Mw: 33821
================
*FHL1_RAT*
accession n°: FHL1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 661 (rattus_soleus-dige_4.5-9.5)
pI: 10.00 Mw: 33821
================
*FHL1_RAT*
accession n°: FHL1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 366 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 366 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 689 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 36168
================
*ANXA4_RAT*
accession n°: ANXA4_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 689 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 36168
================
*ANXA4_RAT*
accession n°: ANXA4_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 183 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 38757
================
*FETUA_RAT*
accession n°: FETUA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 183 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 38757
================
*FETUA_RAT*
accession n°: FETUA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 704 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32931
================
*TPM2_RAT*
accession n°: TPM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 704 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32931
================
*TPM2_RAT*
accession n°: TPM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 149 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 149 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 980 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 980 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 65 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 71047
================
*ACADV_RAT*
accession n°: ACADV_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 65 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 71047
================
*ACADV_RAT*
accession n°: ACADV_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 742 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 742 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 560 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 560 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 477 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 477 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 787 (rattus_soleus-dige_4.5-9.5)
pI: 7.70 Mw: 27345
================
*TPIS_RAT*
accession n°: TPIS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 787 (rattus_soleus-dige_4.5-9.5)
pI: 7.70 Mw: 27345
================
*TPIS_RAT*
accession n°: TPIS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 557 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 557 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 337 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 56966
================
*ALDH2_RAT*
accession n°: ALDH2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 337 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 56966
================
*ALDH2_RAT*
accession n°: ALDH2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 614 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 36117
================
*MDHM_RAT*
accession n°: MDHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 614 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 36117
================
*MDHM_RAT*
accession n°: MDHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 583 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 583 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 19 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 19 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 655 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 30851
================
*VDAC1_RAT*
accession n°: VDAC1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 655 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 30851
================
*VDAC1_RAT*
accession n°: VDAC1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 610 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 610 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 740 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 740 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 862 (rattus_soleus-dige_4.5-9.5)
pI: 9.60 Mw: 24887
================
*SODM_RAT*
accession n°: SODM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 862 (rattus_soleus-dige_4.5-9.5)
pI: 9.60 Mw: 24887
================
*SODM_RAT*
accession n°: SODM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 741 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 741 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 348 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 348 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 284 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 56514
================
*AMPL_RAT*
accession n°: AMPL_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 284 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 56514
================
*AMPL_RAT*
accession n°: AMPL_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 86 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 74564
================
*LMNA_RAT*
accession n°: LMNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 86 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 74564
================
*LMNA_RAT*
accession n°: LMNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 425 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 54714
================
*FUMH_RAT*
accession n°: FUMH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 425 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 54714
================
*FUMH_RAT*
accession n°: FUMH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 626 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 36631
================
*MDHC_RAT*
accession n°: MDHC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 626 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 36631
================
*MDHC_RAT*
accession n°: MDHC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 223 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 223 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 618 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 36117
================
*MDHM_RAT*
accession n°: MDHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 618 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 36117
================
*MDHM_RAT*
accession n°: MDHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 484 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 376261
================
*BRCA2_RAT*
accession n°: BRCA2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 484 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 376261
================
*BRCA2_RAT*
accession n°: BRCA2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 239 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 239 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 104 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 104 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 367 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 367 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 168 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 46419
================
*SPA3L_RAT*
accession n°: SPA3L_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 168 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 46419
================
*SPA3L_RAT*
accession n°: SPA3L_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 89 (rattus_soleus-dige_4.5-9.5)
pI: 8.60 Mw: 52060
================
*HEMO_RAT*
accession n°: HEMO_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 89 (rattus_soleus-dige_4.5-9.5)
pI: 8.60 Mw: 52060
================
*HEMO_RAT*
accession n°: HEMO_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 410 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 410 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 523 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 47683
================
*AATM_RAT*
accession n°: AATM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 523 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 47683
================
*AATM_RAT*
accession n°: AATM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 338 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 338 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 211 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 59955
================
*TCPE_RAT*
accession n°: TCPE_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 211 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 59955
================
*TCPE_RAT*
accession n°: TCPE_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 150 (rattus_soleus-dige_4.5-9.5)
pI: 7.00 Mw: 74634
================
*CPT2_RAT*
accession n°: CPT2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 150 (rattus_soleus-dige_4.5-9.5)
pI: 7.00 Mw: 74634
================
*CPT2_RAT*
accession n°: CPT2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 780 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 780 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 138 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 138 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 133 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 133 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 50 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 80331
================
*NDUS1_RAT*
accession n°: NDUS1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 50 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 80331
================
*NDUS1_RAT*
accession n°: NDUS1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 449 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 48020
================
*SAHH_RAT*
accession n°: SAHH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 449 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 48020
================
*SAHH_RAT*
accession n°: SAHH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 830 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 830 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 871 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 871 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 715 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 27936
================
*GSTO1_RAT*
accession n°: GSTO1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 715 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 27936
================
*GSTO1_RAT*
accession n°: GSTO1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 757 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 24690
================
*HPRT_RAT*
accession n°: HPRT_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 757 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 24690
================
*HPRT_RAT*
accession n°: HPRT_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 612 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 612 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 855 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 18809
================
*ATP5H_RAT*
accession n°: ATP5H_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 855 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 18809
================
*ATP5H_RAT*
accession n°: ATP5H_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 258 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 258 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 811 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 22256
================
*MYL3_RAT*
accession n°: MYL3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 811 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 22256
================
*MYL3_RAT*
accession n°: MYL3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 370 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 370 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 269 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 269 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 615 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 615 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 889 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 17551
================
*HSPB6_RAT*
accession n°: HSPB6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 889 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 17551
================
*HSPB6_RAT*
accession n°: HSPB6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 294 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 53069
================
*SBP1_RAT*
accession n°: SBP1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 294 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 53069
================
*SBP1_RAT*
accession n°: SBP1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 544 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 544 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 846 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 20781
================
*MYL1_RAT*
accession n°: MYL1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 846 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 20781
================
*MYL1_RAT*
accession n°: MYL1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 442 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 442 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 463 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 463 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 750 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 750 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 631 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 36117
================
*MDHM_RAT*
accession n°: MDHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 631 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 36117
================
*MDHM_RAT*
accession n°: MDHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 208 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 208 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 570 (rattus_soleus-dige_4.5-9.5)
pI: 8.50 Mw: 40753
================
*NDUAA_RAT*
accession n°: NDUAA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 570 (rattus_soleus-dige_4.5-9.5)
pI: 8.50 Mw: 40753
================
*NDUAA_RAT*
accession n°: NDUAA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 388 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 51554
================
*GUAD_RAT*
accession n°: GUAD_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 388 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 51554
================
*GUAD_RAT*
accession n°: GUAD_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 312 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 53069
================
*SBP1_RAT*
accession n°: SBP1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 312 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 53069
================
*SBP1_RAT*
accession n°: SBP1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 916 (rattus_soleus-dige_4.5-9.5)
pI: 9.10 Mw: 18749
================
*COF1_RAT*
accession n°: COF1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 916 (rattus_soleus-dige_4.5-9.5)
pI: 9.10 Mw: 18749
================
*COF1_RAT*
accession n°: COF1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 650 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 30851
================
*VDAC1_RAT*
accession n°: VDAC1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 650 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 30851
================
*VDAC1_RAT*
accession n°: VDAC1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 747 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 747 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 261 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 261 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 199 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 199 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 748 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 29326
================
*1433E_RAT*
accession n°: 1433E_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 748 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 29326
================
*1433E_RAT*
accession n°: 1433E_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 164 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 56624
================
*SCOT1_RAT*
accession n°: SCOT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 164 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 56624
================
*SCOT1_RAT*
accession n°: SCOT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 530 (rattus_soleus-dige_4.5-9.5)
pI: 10.30 Mw: 63052
================
*THAP4_RAT*
accession n°: THAP4_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 530 (rattus_soleus-dige_4.5-9.5)
pI: 10.30 Mw: 63052
================
*THAP4_RAT*
accession n°: THAP4_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 157 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 74564
================
*LMNA_RAT*
accession n°: LMNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 157 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 74564
================
*LMNA_RAT*
accession n°: LMNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 424 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 52927
================
*NDUS2_RAT*
accession n°: NDUS2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 424 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 52927
================
*NDUS2_RAT*
accession n°: NDUS2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 978 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 978 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 433 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 433 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 984 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 312623
================
*APC_RAT*
accession n°: APC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 984 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 312623
================
*APC_RAT*
accession n°: APC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 420 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 80331
================
*NDUS1_RAT*
accession n°: NDUS1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 420 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 80331
================
*NDUS1_RAT*
accession n°: NDUS1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 350 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 350 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 938 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 938 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 749 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 15490
================
*HBA_RAT*
accession n°: HBA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 749 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 15490
================
*HBA_RAT*
accession n°: HBA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 87 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 87 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 351 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 53481
================
*DESM_RAT*
accession n°: DESM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 351 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 53481
================
*DESM_RAT*
accession n°: DESM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 693 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 693 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 533 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 62638
================
*DPYL2_RAT*
accession n°: DPYL2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 533 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 62638
================
*DPYL2_RAT*
accession n°: DPYL2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 426 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 54714
================
*FUMH_RAT*
accession n°: FUMH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 426 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 54714
================
*FUMH_RAT*
accession n°: FUMH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 391 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 47811
================
*KCRS_RAT*
accession n°: KCRS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 391 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 47811
================
*KCRS_RAT*
accession n°: KCRS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 796 (rattus_soleus-dige_4.5-9.5)
pI: 7.70 Mw: 27345
================
*TPIS_RAT*
accession n°: TPIS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 796 (rattus_soleus-dige_4.5-9.5)
pI: 7.70 Mw: 27345
================
*TPIS_RAT*
accession n°: TPIS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 505 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 39783
================
*ALDOA_RAT*
accession n°: ALDOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 505 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 39783
================
*ALDOA_RAT*
accession n°: ALDOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 162 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 46419
================
*SPA3L_RAT*
accession n°: SPA3L_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 162 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 46419
================
*SPA3L_RAT*
accession n°: SPA3L_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 144 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 144 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 949 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 949 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 195 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 195 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 42 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 42 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 435 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 47493
================
*PRS6B_RAT*
accession n°: PRS6B_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 435 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 47493
================
*PRS6B_RAT*
accession n°: PRS6B_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 772 (rattus_soleus-dige_4.5-9.5)
pI: 9.30 Mw: 25360
================
*GSTA3_RAT*
accession n°: GSTA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 772 (rattus_soleus-dige_4.5-9.5)
pI: 9.30 Mw: 25360
================
*GSTA3_RAT*
accession n°: GSTA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 120 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 120 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 551 (rattus_soleus-dige_4.5-9.5)
pI: 8.50 Mw: 40753
================
*NDUAA_RAT*
accession n°: NDUAA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 551 (rattus_soleus-dige_4.5-9.5)
pI: 8.50 Mw: 40753
================
*NDUAA_RAT*
accession n°: NDUAA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 860 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 20902
================
*PEBP1_RAT*
accession n°: PEBP1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 860 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 20902
================
*PEBP1_RAT*
accession n°: PEBP1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 849 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 19761
================
*APT_RAT*
accession n°: APT_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 849 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 19761
================
*APT_RAT*
accession n°: APT_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 206 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 57044
================
*PDIA3_RAT*
accession n°: PDIA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 206 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 57044
================
*PDIA3_RAT*
accession n°: PDIA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 738 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 28365
================
*1433F_RAT*
accession n°: 1433F_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 738 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 28365
================
*1433F_RAT*
accession n°: 1433F_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 475 (rattus_soleus-dige_4.5-9.5)
pI: 6.00 Mw: 46060
================
*ACY1A_RAT*
accession n°: ACY1A_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 475 (rattus_soleus-dige_4.5-9.5)
pI: 6.00 Mw: 46060
================
*ACY1A_RAT*
accession n°: ACY1A_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 974 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 974 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 299 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 299 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 957 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 16073
================
*SODC_RAT*
accession n°: SODC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 957 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 16073
================
*SODC_RAT*
accession n°: SODC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 883 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 883 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 119 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 119 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 633 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 29217
================
*TPM3_RAT*
accession n°: TPM3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 633 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 29217
================
*TPM3_RAT*
accession n°: TPM3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 700 (rattus_soleus-dige_4.5-9.5)
pI: 10.00 Mw: 33821
================
*FHL1_RAT*
accession n°: FHL1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 700 (rattus_soleus-dige_4.5-9.5)
pI: 10.00 Mw: 33821
================
*FHL1_RAT*
accession n°: FHL1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 888 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 18868
================
*MLRV_RAT*
accession n°: MLRV_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 888 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 18868
================
*MLRV_RAT*
accession n°: MLRV_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 230 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 230 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 670 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 33205
================
*THTM_RAT*
accession n°: THTM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 670 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 33205
================
*THTM_RAT*
accession n°: THTM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 522 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 47683
================
*AATM_RAT*
accession n°: AATM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 522 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 47683
================
*AATM_RAT*
accession n°: AATM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 461 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 461 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 57 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 57 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 300 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 53069
================
*SBP1_RAT*
accession n°: SBP1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 300 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 53069
================
*SBP1_RAT*
accession n°: SBP1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 222 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 222 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 113 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 72473
================
*GRP78_RAT*
accession n°: GRP78_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 113 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 72473
================
*GRP78_RAT*
accession n°: GRP78_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 409 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 53500
================
*QCR1_RAT*
accession n°: QCR1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 409 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 53500
================
*QCR1_RAT*
accession n°: QCR1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 296 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 296 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 828 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 27343
================
*HCD2_RAT*
accession n°: HCD2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 828 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 27343
================
*HCD2_RAT*
accession n°: HCD2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 578 (rattus_soleus-dige_4.5-9.5)
pI: 6.50 Mw: 40044
================
*IDH3A_RAT*
accession n°: IDH3A_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 578 (rattus_soleus-dige_4.5-9.5)
pI: 6.50 Mw: 40044
================
*IDH3A_RAT*
accession n°: IDH3A_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 731 (rattus_soleus-dige_4.5-9.5)
pI: 9.10 Mw: 30250
================
*PSB7_RAT*
accession n°: PSB7_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 731 (rattus_soleus-dige_4.5-9.5)
pI: 9.10 Mw: 30250
================
*PSB7_RAT*
accession n°: PSB7_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 448 (rattus_soleus-dige_4.5-9.5)
pI: 9.40 Mw: 43883
================
*ODPA_RAT*
accession n°: ODPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 448 (rattus_soleus-dige_4.5-9.5)
pI: 9.40 Mw: 43883
================
*ODPA_RAT*
accession n°: ODPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 504 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 39783
================
*ALDOA_RAT*
accession n°: ALDOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 504 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 39783
================
*ALDOA_RAT*
accession n°: ALDOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 257 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 50095
================
*TBB5_RAT*
accession n°: TBB5_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 257 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 50095
================
*TBB5_RAT*
accession n°: TBB5_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 885 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 18868
================
*MLRV_RAT*
accession n°: MLRV_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 885 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 18868
================
*MLRV_RAT*
accession n°: MLRV_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 956 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 16073
================
*SODC_RAT*
accession n°: SODC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 956 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 16073
================
*SODC_RAT*
accession n°: SODC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 640 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 640 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 173 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 173 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 762 (rattus_soleus-dige_4.5-9.5)
pI: 9.40 Mw: 31895
================
*ECHM_RAT*
accession n°: ECHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 762 (rattus_soleus-dige_4.5-9.5)
pI: 9.40 Mw: 31895
================
*ECHM_RAT*
accession n°: ECHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 638 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 36631
================
*MDHC_RAT*
accession n°: MDHC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 638 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 36631
================
*MDHC_RAT*
accession n°: MDHC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 904 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 17551
================
*HSPB6_RAT*
accession n°: HSPB6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 904 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 17551
================
*HSPB6_RAT*
accession n°: HSPB6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 90 (rattus_soleus-dige_4.5-9.5)
pI: 8.60 Mw: 52060
================
*HEMO_RAT*
accession n°: HEMO_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 90 (rattus_soleus-dige_4.5-9.5)
pI: 8.60 Mw: 52060
================
*HEMO_RAT*
accession n°: HEMO_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 450 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 48020
================
*SAHH_RAT*
accession n°: SAHH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 450 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 48020
================
*SAHH_RAT*
accession n°: SAHH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 584 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 584 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 635 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 39299
================
*ODPB_RAT*
accession n°: ODPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 635 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 39299
================
*ODPB_RAT*
accession n°: ODPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 274 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 274 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 169 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 38757
================
*FETUA_RAT*
accession n°: FETUA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 169 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 38757
================
*FETUA_RAT*
accession n°: FETUA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 596 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 596 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 429 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 53500
================
*QCR1_RAT*
accession n°: QCR1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 429 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 53500
================
*QCR1_RAT*
accession n°: QCR1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 688 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 688 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 651 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 651 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 336 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 53116
================
*CNDP2_RAT*
accession n°: CNDP2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 336 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 53116
================
*CNDP2_RAT*
accession n°: CNDP2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 907 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 17551
================
*HSPB6_RAT*
accession n°: HSPB6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 907 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 17551
================
*HSPB6_RAT*
accession n°: HSPB6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 783 (rattus_soleus-dige_4.5-9.5)
pI: 9.30 Mw: 25360
================
*GSTA3_RAT*
accession n°: GSTA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 783 (rattus_soleus-dige_4.5-9.5)
pI: 9.30 Mw: 25360
================
*GSTA3_RAT*
accession n°: GSTA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 381 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 381 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 829 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 27343
================
*HCD2_RAT*
accession n°: HCD2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 829 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 27343
================
*HCD2_RAT*
accession n°: HCD2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 760 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 760 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 488 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 488 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 636 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 36631
================
*MDHC_RAT*
accession n°: MDHC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 636 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 36631
================
*MDHC_RAT*
accession n°: MDHC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 115 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 72473
================
*GRP78_RAT*
accession n°: GRP78_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 115 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 72473
================
*GRP78_RAT*
accession n°: GRP78_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 745 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 745 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 94 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 74564
================
*LMNA_RAT*
accession n°: LMNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 94 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 74564
================
*LMNA_RAT*
accession n°: LMNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 139 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 46760
================
*SPA3K_RAT*
accession n°: SPA3K_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 139 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 46760
================
*SPA3K_RAT*
accession n°: SPA3K_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 774 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 774 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 432 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 432 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 494 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 494 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 153 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 46760
================
*SPA3K_RAT*
accession n°: SPA3K_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 153 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 46760
================
*SPA3K_RAT*
accession n°: SPA3K_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 999 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 53757
================
*VIME_RAT*
accession n°: VIME_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 999 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 53757
================
*VIME_RAT*
accession n°: VIME_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 825 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 24860
================
*PRDX6_RAT*
accession n°: PRDX6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 825 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 24860
================
*PRDX6_RAT*
accession n°: PRDX6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 379 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 379 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 137 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 137 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 213 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 213 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 202 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 57044
================
*PDIA3_RAT*
accession n°: PDIA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 202 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 57044
================
*PDIA3_RAT*
accession n°: PDIA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 459 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 53757
================
*VIME_RAT*
accession n°: VIME_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 459 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 53757
================
*VIME_RAT*
accession n°: VIME_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 735 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 27925
================
*1433Z_RAT*
accession n°: 1433Z_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 735 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 27925
================
*1433Z_RAT*
accession n°: 1433Z_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 673 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 36168
================
*ANXA4_RAT*
accession n°: ANXA4_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 673 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 36168
================
*ANXA4_RAT*
accession n°: ANXA4_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 92 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 92 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 492 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 492 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 397 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 397 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 114 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 46760
================
*SPA3K_RAT*
accession n°: SPA3K_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 114 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 46760
================
*SPA3K_RAT*
accession n°: SPA3K_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 220 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 57044
================
*PDIA3_RAT*
accession n°: PDIA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 220 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 57044
================
*PDIA3_RAT*
accession n°: PDIA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 85 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 56699
================
*K1C10_RAT*
accession n°: K1C10_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 85 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 56699
================
*K1C10_RAT*
accession n°: K1C10_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 486 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 46862
================
*IVD_RAT*
accession n°: IVD_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 486 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 46862
================
*IVD_RAT*
accession n°: IVD_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 423 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 54714
================
*FUMH_RAT*
accession n°: FUMH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 423 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 54714
================
*FUMH_RAT*
accession n°: FUMH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 914 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 18868
================
*MLRV_RAT*
accession n°: MLRV_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 914 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 18868
================
*MLRV_RAT*
accession n°: MLRV_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 589 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32718
================
*TPM1_RAT*
accession n°: TPM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 589 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32718
================
*TPM1_RAT*
accession n°: TPM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 439 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 439 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 276 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 276 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 571 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 52927
================
*NDUS2_RAT*
accession n°: NDUS2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 571 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 52927
================
*NDUS2_RAT*
accession n°: NDUS2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 714 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 29859
================
*PHB_RAT*
accession n°: PHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 714 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 29859
================
*PHB_RAT*
accession n°: PHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 864 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 21941
================
*PRDX2_RAT*
accession n°: PRDX2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 864 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 21941
================
*PRDX2_RAT*
accession n°: PRDX2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 518 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 312623
================
*APC_RAT*
accession n°: APC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 518 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 312623
================
*APC_RAT*
accession n°: APC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 977 (rattus_soleus-dige_4.5-9.5)
pI: 6.40 Mw: 20190
================
*PARK7_RAT*
accession n°: PARK7_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 977 (rattus_soleus-dige_4.5-9.5)
pI: 6.40 Mw: 20190
================
*PARK7_RAT*
accession n°: PARK7_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 62 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 62 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 823 (rattus_soleus-dige_4.5-9.5)
pI: 4.30 Mw: 17135
================
*MYL6_RAT*
accession n°: MYL6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 823 (rattus_soleus-dige_4.5-9.5)
pI: 4.30 Mw: 17135
================
*MYL6_RAT*
accession n°: MYL6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 803 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 24860
================
*PRDX6_RAT*
accession n°: PRDX6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 803 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 24860
================
*PRDX6_RAT*
accession n°: PRDX6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 645 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 645 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 476 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 476 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 553 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 553 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 586 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 36230
================
*ALDR_RAT*
accession n°: ALDR_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 586 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 36230
================
*ALDR_RAT*
accession n°: ALDR_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 375 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 48542
================
*PDIA6_RAT*
accession n°: PDIA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 375 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 48542
================
*PDIA6_RAT*
accession n°: PDIA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 945 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 17386
================
*NDKB_RAT*
accession n°: NDKB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 945 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 17386
================
*NDKB_RAT*
accession n°: NDKB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 140 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 140 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 761 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 761 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 665 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 35779
================
*ANXA5_RAT*
accession n°: ANXA5_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 665 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 35779
================
*ANXA5_RAT*
accession n°: ANXA5_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 798 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 22936
================
*HSPB1_RAT*
accession n°: HSPB1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 798 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 22936
================
*HSPB1_RAT*
accession n°: HSPB1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 127 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 127 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 503 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 39783
================
*ALDOA_RAT*
accession n°: ALDOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 503 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 39783
================
*ALDOA_RAT*
accession n°: ALDOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 210 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 57044
================
*PDIA3_RAT*
accession n°: PDIA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 210 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 57044
================
*PDIA3_RAT*
accession n°: PDIA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 436 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 436 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 431 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 431 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 861 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 18809
================
*ATP5H_RAT*
accession n°: ATP5H_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 861 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 18809
================
*ATP5H_RAT*
accession n°: ATP5H_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 123 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 46760
================
*SPA3K_RAT*
accession n°: SPA3K_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 123 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 46760
================
*SPA3K_RAT*
accession n°: SPA3K_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 621 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 621 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 88 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 74097
================
*GRP75_RAT*
accession n°: GRP75_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 88 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 74097
================
*GRP75_RAT*
accession n°: GRP75_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 809 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 22936
================
*HSPB1_RAT*
accession n°: HSPB1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 809 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 22936
================
*HSPB1_RAT*
accession n°: HSPB1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 319 (rattus_soleus-dige_4.5-9.5)
pI: 6.70 Mw: 54530
================
*AL9A1_RAT*
accession n°: AL9A1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 319 (rattus_soleus-dige_4.5-9.5)
pI: 6.70 Mw: 54530
================
*AL9A1_RAT*
accession n°: AL9A1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 345 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 345 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 178 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 61179
================
*TCPG_RAT*
accession n°: TCPG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 178 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 61179
================
*TCPG_RAT*
accession n°: TCPG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 281 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 281 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 238 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 238 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 561 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32931
================
*TPM2_RAT*
accession n°: TPM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 561 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32931
================
*TPM2_RAT*
accession n°: TPM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 744 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 744 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 341 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 341 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 992 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 32607
================
*DHSB_RAT*
accession n°: DHSB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 992 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 32607
================
*DHSB_RAT*
accession n°: DHSB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 754 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 754 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 28908
================
*PGAM2_RAT*
accession n°: PGAM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 358 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 51554
================
*GUAD_RAT*
accession n°: GUAD_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 358 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 51554
================
*GUAD_RAT*
accession n°: GUAD_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 996 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 996 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 42366
================
*ACTS_RAT*
accession n°: ACTS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 917 (rattus_soleus-dige_4.5-9.5)
pI: 9.10 Mw: 18749
================
*COF1_RAT*
accession n°: COF1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 917 (rattus_soleus-dige_4.5-9.5)
pI: 9.10 Mw: 18749
================
*COF1_RAT*
accession n°: COF1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 21 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 67868
================
*MOES_RAT*
accession n°: MOES_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 21 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 67868
================
*MOES_RAT*
accession n°: MOES_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 143 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 70427
================
*HSP71_RAT*
accession n°: HSP71_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 143 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 70427
================
*HSP71_RAT*
accession n°: HSP71_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 646 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 646 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 415 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 51653
================
*RINI_RAT*
accession n°: RINI_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 415 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 51653
================
*RINI_RAT*
accession n°: RINI_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 419 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 419 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 788 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 24690
================
*HPRT_RAT*
accession n°: HPRT_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 788 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 24690
================
*HPRT_RAT*
accession n°: HPRT_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 753 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 753 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 536 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 536 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 676 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 33205
================
*THTM_RAT*
accession n°: THTM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 676 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 33205
================
*THTM_RAT*
accession n°: THTM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 253 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 50225
================
*TBB2C_RAT*
accession n°: TBB2C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 253 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 50225
================
*TBB2C_RAT*
accession n°: TBB2C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 405 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 405 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 155 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 155 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 532 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32931
================
*TPM2_RAT*
accession n°: TPM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 532 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32931
================
*TPM2_RAT*
accession n°: TPM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 134 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 134 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 677 (rattus_soleus-dige_4.5-9.5)
pI: 9.60 Mw: 35679
================
*3HIDH_RAT*
accession n°: 3HIDH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 677 (rattus_soleus-dige_4.5-9.5)
pI: 9.60 Mw: 35679
================
*3HIDH_RAT*
accession n°: 3HIDH_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 234 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 234 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 61088
================
*CH60_RAT*
accession n°: CH60_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 411 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 411 (rattus_soleus-dige_4.5-9.5)
pI: 5.00 Mw: 41095
================
*NDRG2_RAT*
accession n°: NDRG2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 95 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 95 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 763 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 22936
================
*HSPB1_RAT*
accession n°: HSPB1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 763 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 22936
================
*HSPB1_RAT*
accession n°: HSPB1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 154 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 154 (rattus_soleus-dige_4.5-9.5)
pI: 5.30 Mw: 76106
================
*ANXA6_RAT*
accession n°: ANXA6_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 755 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 755 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 460 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 460 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 597 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 597 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 36874
================
*LDHB_RAT*
accession n°: LDHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 109 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 109 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 976 (rattus_soleus-dige_4.5-9.5)
pI: 8.00 Mw: 28563
================
*PRDX3_RAT*
accession n°: PRDX3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 976 (rattus_soleus-dige_4.5-9.5)
pI: 8.00 Mw: 28563
================
*PRDX3_RAT*
accession n°: PRDX3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 873 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 873 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 936 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 17049
================
*IF5A1_RAT*
accession n°: IF5A1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 936 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 17049
================
*IF5A1_RAT*
accession n°: IF5A1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 769 (rattus_soleus-dige_4.5-9.5)
pI: 9.30 Mw: 25360
================
*GSTA3_RAT*
accession n°: GSTA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 769 (rattus_soleus-dige_4.5-9.5)
pI: 9.30 Mw: 25360
================
*GSTA3_RAT*
accession n°: GSTA3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 605 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 36117
================
*MDHM_RAT*
accession n°: MDHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 605 (rattus_soleus-dige_4.5-9.5)
pI: 9.80 Mw: 36117
================
*MDHM_RAT*
accession n°: MDHM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 826 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 30100
================
*APOA1_RAT*
accession n°: APOA1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 826 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 30100
================
*APOA1_RAT*
accession n°: APOA1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 500 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 500 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 70 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 83571
================
*HS90B_RAT*
accession n°: HS90B_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 70 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 83571
================
*HS90B_RAT*
accession n°: HS90B_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 535 (rattus_soleus-dige_4.5-9.5)
pI: 8.50 Mw: 40753
================
*NDUAA_RAT*
accession n°: NDUAA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 535 (rattus_soleus-dige_4.5-9.5)
pI: 8.50 Mw: 40753
================
*NDUAA_RAT*
accession n°: NDUAA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 156 (rattus_soleus-dige_4.5-9.5)
pI: 6.00 Mw: 57546
================
*SYDC_RAT*
accession n°: SYDC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 156 (rattus_soleus-dige_4.5-9.5)
pI: 6.00 Mw: 57546
================
*SYDC_RAT*
accession n°: SYDC_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 743 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 743 (rattus_soleus-dige_4.5-9.5)
pI: 7.10 Mw: 29698
================
*CAH3_RAT*
accession n°: CAH3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 604 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 604 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 950 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 950 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 179 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 179 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 498 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 498 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 792 (rattus_soleus-dige_4.5-9.5)
pI: 7.70 Mw: 27345
================
*TPIS_RAT*
accession n°: TPIS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 792 (rattus_soleus-dige_4.5-9.5)
pI: 7.70 Mw: 27345
================
*TPIS_RAT*
accession n°: TPIS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 248 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 57315
================
*PDIA1_RAT*
accession n°: PDIA1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 248 (rattus_soleus-dige_4.5-9.5)
pI: 4.70 Mw: 57315
================
*PDIA1_RAT*
accession n°: PDIA1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 165 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 67637
================
*ODP2_RAT*
accession n°: ODP2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 165 (rattus_soleus-dige_4.5-9.5)
pI: 9.50 Mw: 67637
================
*ODP2_RAT*
accession n°: ODP2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 648 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 32115
================
*PIPNA_RAT*
accession n°: PIPNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 648 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 32115
================
*PIPNA_RAT*
accession n°: PIPNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 76 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 83571
================
*HS90B_RAT*
accession n°: HS90B_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 76 (rattus_soleus-dige_4.5-9.5)
pI: 4.80 Mw: 83571
================
*HS90B_RAT*
accession n°: HS90B_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 91 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 91 (rattus_soleus-dige_4.5-9.5)
pI: 6.80 Mw: 72596
================
*DHSA_RAT*
accession n°: DHSA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 493 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 493 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 148 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 70427
================
*HSP71_RAT*
accession n°: HSP71_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 148 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 70427
================
*HSP71_RAT*
accession n°: HSP71_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 313 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 313 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 53767
================
*TRI72_RAT*
accession n°: TRI72_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 32 (rattus_soleus-dige_4.5-9.5)
pI: 8.70 Mw: 86121
================
*ACON_RAT*
accession n°: ACON_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 32 (rattus_soleus-dige_4.5-9.5)
pI: 8.70 Mw: 86121
================
*ACON_RAT*
accession n°: ACON_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 334 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 334 (rattus_soleus-dige_4.5-9.5)
pI: 9.70 Mw: 59831
================
*ATPA_RAT*
accession n°: ATPA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 539 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 46419
================
*SPA3L_RAT*
accession n°: SPA3L_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 539 (rattus_soleus-dige_4.5-9.5)
pI: 5.40 Mw: 46419
================
*SPA3L_RAT*
accession n°: SPA3L_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 981 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 981 (rattus_soleus-dige_4.5-9.5)
pI: 9.00 Mw: 36090
================
*G3P_RAT*
accession n°: G3P_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 394 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 394 (rattus_soleus-dige_4.5-9.5)
pI: 7.80 Mw: 47326
================
*ENOB_RAT*
accession n°: ENOB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 975 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 975 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 17203
================
*MYG_RAT*
accession n°: MYG_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 329 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 74564
================
*LMNA_RAT*
accession n°: LMNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 329 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 74564
================
*LMNA_RAT*
accession n°: LMNA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 515 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 515 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 608 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 608 (rattus_soleus-dige_4.5-9.5)
pI: 5.80 Mw: 31196
================
*TNNT1_RAT*
accession n°: TNNT1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 507 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 39783
================
*ALDOA_RAT*
accession n°: ALDOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 507 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 39783
================
*ALDOA_RAT*
accession n°: ALDOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 730 (rattus_soleus-dige_4.5-9.5)
pI: 5.70 Mw: 28730
================
*PSME1_RAT*
accession n°: PSME1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 730 (rattus_soleus-dige_4.5-9.5)
pI: 5.70 Mw: 28730
================
*PSME1_RAT*
accession n°: PSME1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 347 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 347 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 56318
================
*ATPB_RAT*
accession n°: ATPB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 84 (rattus_soleus-dige_4.5-9.5)
pI: 8.60 Mw: 52060
================
*HEMO_RAT*
accession n°: HEMO_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 84 (rattus_soleus-dige_4.5-9.5)
pI: 8.60 Mw: 52060
================
*HEMO_RAT*
accession n°: HEMO_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 491 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 376261
================
*BRCA2_RAT*
accession n°: BRCA2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 491 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 376261
================
*BRCA2_RAT*
accession n°: BRCA2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 872 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 872 (rattus_soleus-dige_4.5-9.5)
pI: 6.90 Mw: 20076
================
*CRYAB_RAT*
accession n°: CRYAB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 182 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 182 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 320 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 320 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 581 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 581 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 710 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 29859
================
*PHB_RAT*
accession n°: PHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 710 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 29859
================
*PHB_RAT*
accession n°: PHB_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 107 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 74097
================
*GRP75_RAT*
accession n°: GRP75_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 107 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 74097
================
*GRP75_RAT*
accession n°: GRP75_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 326 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 59219
================
*K2C75_RAT*
accession n°: K2C75_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 326 (rattus_soleus-dige_4.5-9.5)
pI: 8.90 Mw: 59219
================
*K2C75_RAT*
accession n°: K2C75_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 746 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 27445
================
*6PGL_RAT*
accession n°: 6PGL_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 746 (rattus_soleus-dige_4.5-9.5)
pI: 5.50 Mw: 27445
================
*6PGL_RAT*
accession n°: 6PGL_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 54 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 53481
================
*DESM_RAT*
accession n°: DESM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 54 (rattus_soleus-dige_4.5-9.5)
pI: 5.10 Mw: 53481
================
*DESM_RAT*
accession n°: DESM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 135 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 135 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 70682
================
*ALBU_RAT*
accession n°: ALBU_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 541 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 541 (rattus_soleus-dige_4.5-9.5)
pI: 6.60 Mw: 43246
================
*KCRM_RAT*
accession n°: KCRM_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 789 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 22936
================
*HSPB1_RAT*
accession n°: HSPB1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 789 (rattus_soleus-dige_4.5-9.5)
pI: 6.10 Mw: 22936
================
*HSPB1_RAT*
accession n°: HSPB1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 147 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 147 (rattus_soleus-dige_4.5-9.5)
pI: 6.30 Mw: 61650
================
*PGM1_RAT*
accession n°: PGM1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 52 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 80331
================
*NDUS1_RAT*
accession n°: NDUS1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 52 (rattus_soleus-dige_4.5-9.5)
pI: 5.60 Mw: 80331
================
*NDUS1_RAT*
accession n°: NDUS1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 705 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32931
================
*TPM2_RAT*
accession n°: TPM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 705 (rattus_soleus-dige_4.5-9.5)
pI: 4.50 Mw: 32931
================
*TPM2_RAT*
accession n°: TPM2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 340 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 340 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 915 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 19071
================
*MLRS_RAT*
accession n°: MLRS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 915 (rattus_soleus-dige_4.5-9.5)
pI: 4.60 Mw: 19071
================
*MLRS_RAT*
accession n°: MLRS_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 385 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 30851
================
*VDAC1_RAT*
accession n°: VDAC1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 385 (rattus_soleus-dige_4.5-9.5)
pI: 9.20 Mw: 30851
================
*VDAC1_RAT*
accession n°: VDAC1_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 817 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 22256
================
*MYL3_RAT*
accession n°: MYL3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 817 (rattus_soleus-dige_4.5-9.5)
pI: 4.90 Mw: 22256
================
*MYL3_RAT*
accession n°: MYL3_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 185 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 62638
================
*DPYL2_RAT*
accession n°: DPYL2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 185 (rattus_soleus-dige_4.5-9.5)
pI: 5.90 Mw: 62638
================
*DPYL2_RAT*
accession n°: DPYL2_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 53 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 53 (rattus_soleus-dige_4.5-9.5)
pI: 5.20 Mw: 71055
================
*HSP7C_RAT*
accession n°: HSP7C_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
[Click to access protein entry]
Spot: 373 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
Spot: 373 (rattus_soleus-dige_4.5-9.5)
pI: 6.20 Mw: 47440
================
*ENOA_RAT*
accession n°: ENOA_RAT
Identification Methods:
* % COVERAGE, MOWSE, NAME
World-2DPAGE Repository Viewer (0069)
Home page of the World-2DPAGE Repository
|
Back to the |
-Get more information by dragging your mouse pointer over any highlighted spot -Click on a highlighted spot to access all its associated protein entries
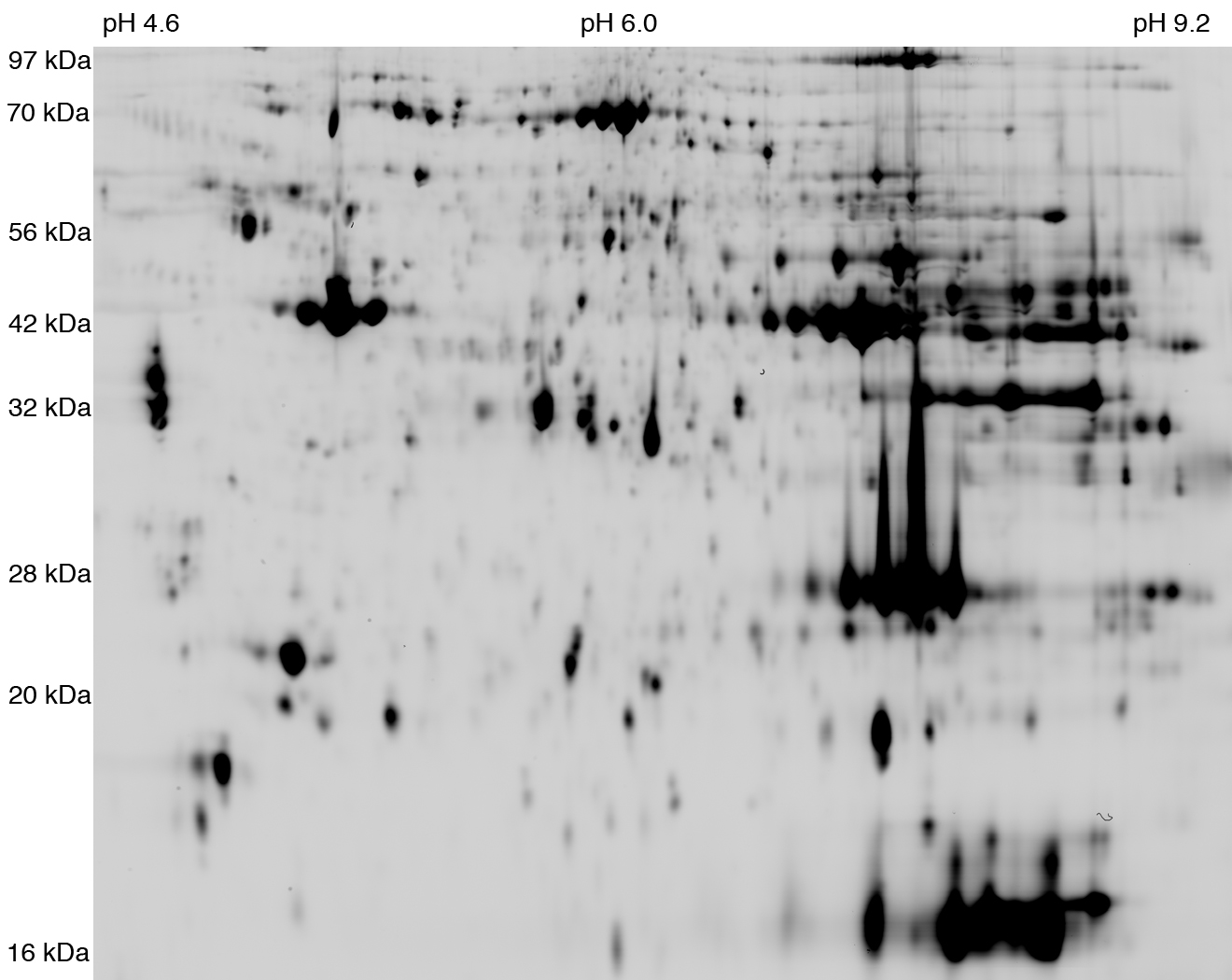
Back to the