|
|
|
|
|
Sample Preparation and Post-separation Analysis
| Searching in 'SWISS-2DPAGE' for entry matching: P0ABK5 |
SWISS-2DPAGE: P0ABK5
P0ABK5:
Nice View - a user-friendly view of this entry
ID CYSK_ECOLI; STANDARD; 2DG.
AC P0ABK5; P11096;
DT 01-AUG-1995, integrated into SWISS-2DPAGE (release 2).
DT 31-MAR-2004, 2D annotation version 5.
DT 19-MAY-2011, general annotation version 13.
DE RecName: Full=Cysteine synthase A; Short=CSase A; EC=2.5.1.47; AltName:
DE Full=O-acetylserine (thiol)-lyase A; Short=OAS-TL A; AltName:
DE Full=O-acetylserine sulfhydrylase A; AltName: Full=Sulfate
DE starvation-induced protein 5; Short=SSI5;.
GN Name=cysK; Synonyms=cysZ; OrderedLocusNames=b2414, JW2407;
OS Escherichia coli.
OC Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales;
OC Enterobacteriaceae; Escherichia.
OX NCBI_TaxID=562;
MT ECOLI, ECOLI4-5, ECOLI4.5-5.5, ECOLI5-6, ECOLI-DIGE4.5-6.5.
IM ECOLI, ECOLI4-5, ECOLI4.5-5.5, ECOLI5-6, ECOLI-DIGE4.5-6.5.
RN [1]
RP MAPPING ON GEL.
RX MEDLINE=96314059; PubMed=8740179; [NCBI, Expasy, EBI, Israel, Japan]
RA Pasquali C., Frutiger S., Wilkins M.R., Hughes G.J., Appel R.D., Bairoch
RA A., Schaller D., Sanchez J.-C., Hochstrasser D.F.;
RT ''''''Two-dimensional gel electrophoresis of Escherichia coli
RT homogenates: the Escherichia coli SWISS-2DPAGE database'';'';'';
RL Electrophoresis 17(1):547-555(1996).
RN [2]
RP MAPPING ON GEL.
RA Vanbogelen R.A., Abshire K.Z., Pertsemlidis A., Clark R.L., Neidhardt F.C.;
RT ''''''Gene-protein database of Escherichia coli K-12, edition 6'';'';'';
RL (IN) Neidhardt et al. (eds.);
RL Escherichia coli and Salmonella: Cellular and Molecular Biology (2nd
RL ed.), pp.2067-2117, ASM Press, Washington DC (1996).
RN [3]
RP MAPPING ON GEL.
RX PubMed=11680886; [NCBI, Expasy, EBI, Israel, Japan]
RA Tonella L., Hoogland C., Binz P.-A., Appel R.D., Hochstrasser D.F.,
RA Sanchez J.-C.;
RT ''''''New perspectives in the Escherichia coli proteome investigation'';'';'';
RL Proteomics 1(1):409-423(2001).
RN [4]
RP MAPPING ON GEL.
RX PubMed=12469338; [NCBI, Expasy, EBI, Israel, Japan]
RA Yan J.X., Devenish A.T., Wait R., Stone T., Lewis S., Fowler S.;
RT ''''''Fluorescence 2-D difference gel electrophoresis and mass
RT spectrometry based proteomic analysis of E. coli'';'';'';
RL Proteomics 2(1):1682-1698(2002).
CC -!- SUBUNIT: HOMODIMER
2D -!- MASTER: ECOLI;
2D -!- PI/MW: SPOT 2D-000L5H=5.80/36099;
2D -!- MAPPING: AMINO ACID COMPOSITION AND MICROSEQUENCE ANALYSIS [1] AND
2D IDENTIFIED ON 2-D GELS BY VANBOGELEN [2].
2D -!- MASTER: ECOLI4-5;
2D -!- PI/MW: SPOT 2D-001HLC=5.01/26299; !FRAGMENT!
2D -!- PI/MW: SPOT 2D-001HLH=4.95/25617; !FRAGMENT!
2D -!- PI/MW: SPOT 2D-001HLU=4.95/24067; !FRAGMENT!
2D -!- PI/MW: SPOT 2D-001HM3=4.93/22242; !FRAGMENT!
2D -!- PEPTIDE MASSES: SPOT 2D-001HLC: 1210.533; 1283.729; 1473.808;
2D 1618.813; 1626.891; 2141.093; 3235.617; 3367.823; TRYPSIN.
2D -!- PEPTIDE MASSES: SPOT 2D-001HLH: 1037.485; 1186.555; 1210.541;
2D 1256.682; 1283.73; 1473.805; 1618.816; 1626.913; 2141.118; 2257.262;
2D 3235.652; TRYPSIN.
2D -!- PEPTIDE MASSES: SPOT 2D-001HLU: 1210.568; 1256.713; 1283.76;
2D 1473.841; 1618.859; 1626.927; 2117.18; 2141.142; 2257.298; 3235.678;
2D 3367.89; TRYPSIN.
2D -!- PEPTIDE MASSES: SPOT 2D-001HM3: 1210.556; 1283.749; 1473.817;
2D 1618.845; 1626.928; 2117.189; 2141.107; 2257.253; 3235.628; TRYPSIN.
2D -!- MAPPING: Peptide mass fingerprinting [3]; SPOT 2D-001HLU:
2D MICROSEQUENCING (XLGA) [3].
2D -!- MASTER: ECOLI4.5-5.5;
2D -!- PI/MW: SPOT 2D-00156W=5.32/32863;
2D -!- PI/MW: SPOT 2D-00158P=5.06/29467;
2D -!- PI/MW: SPOT 2D-00159J=5.04/28314;
2D -!- PI/MW: SPOT 2D-00159M=4.95/28177;
2D -!- PEPTIDE MASSES: SPOT 2D-00156W: 1210.587; 1256.678; 1283.732;
2D 1473.797; 1553.721; 1618.836; 1626.938; 2117.119; 2141.128; 2753.56;
2D 3235.453; TRYPSIN.
2D -!- PEPTIDE MASSES: SPOT 2D-00158P: 1282.98; 1472.97; 1617.97; 1626.08;
2D 2140.12; TRYPSIN.
2D -!- PEPTIDE MASSES: SPOT 2D-00159J: 1210.631; 1256.778; 1283.795;
2D 1473.858; 1618.908; 1626.972; 2141.175; 3235.419; TRYPSIN.
2D -!- PEPTIDE MASSES: SPOT 2D-00159M: 1210.528; 1256.655; 1283.698;
2D 1473.752; 1618.792; 1626.886; 2117.176; 2141.103; TRYPSIN.
2D -!- MAPPING: Peptide mass fingerprinting [3]; SPOT 2D-00158P:
2D MICROSEQUENCING (XYKL) [3].
2D -!- MASTER: ECOLI5-6;
2D -!- PI/MW: SPOT 2D-001L9C=5.78/42978;
2D -!- PI/MW: SPOT 2D-001L9U=5.10/41867;
2D -!- PEPTIDE MASSES: SPOT 2D-001L9C: 1210.565; 1256.722; 1283.75;
2D 1473.811; 1521.782; 1537.771; 1553.783; 1618.861; 1626.926; 1811.995;
2D 2141.145; 2273.216; 2753.494; TRYPSIN.
2D -!- PEPTIDE MASSES: SPOT 2D-001L9U: 1186.634; 1210.589; 1247.641;
2D 1256.749; 1263.691; 1283.787; 1403.776; 1473.851; 1521.834; 1537.798;
2D 1553.816; 1618.9; 1626.976; 2141.155; 2257.293; 2273.278; 2753.565;
2D 3367.848; TRYPSIN.
2D -!- MAPPING: Peptide mass fingerprinting [3].
2D -!- MASTER: ECOLI-DIGE4.5-6.5;
2D -!- PI/MW: SPOT 2D-001WPI=5.83/34342;
2D -!- PEPTIDE MASSES: SPOT 2D-001WPI: 1186.5; 1210.48; 1256.64; 1283.67;
2D 1403.64; 1473.71; 1618.73; 1626.8; 1811.86; 1928.96; 2116.95; 2140.97;
2D 2257.1; 2753.31; 3182.41; 3235.26; TRYPSIN.
2D -!- PEPTIDE SEQUENCES: SPOT 2D-001WPI:
2D (R)GVLKPGVELVEPTSGNTGIALAYVAAAR(G),56-83; (K)ALGANLVLTEGAK(G),105-117;
2D (K)AEEIVASNPEK(Y),126-136; (K)YLLLQQFSNPANPEIHEK(T),137-154;
2D (K)IQGIGAGFIPANLDLK(L),226-241; (K)LQEDESFTNK(N),283-292;
2D (K)NIVVILPSSGER(Y),293-304; (R)YLSTALFADLFTEK(E),305-318.
2D -!- MAPPING: Peptide mass fingerprinting and tandem mass spectrometry [4].
CC ---------------------------------------------------------------------------
CC This SWISS-2DPAGE entry is copyright the Swiss Institute of Bioinformatics.
CC There are no restrictions on its use by non-profit institutions as long as
CC its content is in no way modified and this statement is not removed.
CC Usage by and for commercial entities requires a license agreement (See
CC http://world-2dpage.expasy.org/swiss-2dpage/docs/license.html or send email from legal@sib.swiss).
CC ---------------------------------------------------------------------------
DR 2DBase-Ecoli; P0ABK5; CYSK_ECOLI.
DR UniProtKB/Swiss-Prot; P0ABK5; CYSK_ECOLI.
//
The following 2-D maps are available for this protein:
|
Escherichia coli
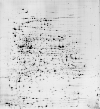 |
Escherichia coli(4-5)
 |
Escherichia coli(4.5-5.5)
 |
|---|
Escherichia coli(5-6)
 |
Escherichia coli DIGE (4.5-6.5)
 |
|
SWISS-2DPAGE (search AC)
|