|
|
|
|
|
Sample Preparation and Post-separation Analysis
| Searching in 'SWISS-2DPAGE' for entry matching: PA2G4_HUMAN |
SWISS-2DPAGE: PA2G4_HUMAN
PA2G4_HUMAN:
Nice View - a user-friendly view of this entry
ID PA2G4_HUMAN; STANDARD; 2DG.
AC Q9UQ80;
DT 09-NOV-2001, integrated into SWISS-2DPAGE (release 15).
DT 15-MAY-2003, 2D annotation version 2.
DT 19-MAY-2011, general annotation version 7.
DE RecName: Full=Proliferation-associated protein 2G4; AltName: Full=Cell
DE cycle protein p38-2G4 homolog; Short=hG4-1; AltName: Full=ErbB3-binding
DE protein 1;.
GN Name=PA2G4; Synonyms=EBP1;
OS Homo sapiens (Human).
OC Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
OC Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini; Catarrhini;
OC Hominidae; Homo.
OX NCBI_TaxID=9606;
MT NUCLEOLI_HELA_1D_HUMAN, NUCLEOLI_HELA_2D_HUMAN.
IM NUCLEOLI_HELA_1D_HUMAN, NUCLEOLI_HELA_2D_HUMAN.
RN [1]
RP MAPPING ON GEL.
RX PubMed=12429849; [NCBI, Expasy, EBI, Israel, Japan]
RA Scherl A., Coute Y., Deon C., Calle A., Kindbeiter K., Sanchez J.-C.,
RA Greco A., Hochstrasser D.F., Diaz J.-J.;
RT ''''''Functional proteomic analysis of human nucleolus'';'';'';
RL Mol. Biol. Cell. 13(1):4100-4109(2002).
1D -!- MASTER: NUCLEOLI_HELA_1D_HUMAN;
1D -!- MW: BAND 1D-001V8R=45729;
1D -!- PEPTIDE SEQUENCES: BAND 1D-001V8R: (R)LVKPGNQNTQVTEAWNK(V),156-172; (R)ITSGPFEPDLYK(S),333-344.
1D -!- MAPPING: Tandem mass spectrometry [1].
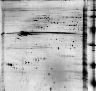
2D -!- MASTER: NUCLEOLI_HELA_2D_HUMAN;
2D -!- PI/MW: SPOT 2D-001VMW=6.43/47236;
2D -!- PI/MW: SPOT 2D-001VN0=6.64/47059;
2D -!- PEPTIDE MASSES: SPOT 2D-001VMW: 1182.4; 1253.4; 1285.5; 1366.5;
2D 1375.6; 1629.7; 2185.1; 2264.1; 2348.2; 2476.3; 2541.3; 3236; TRYPSIN.
2D -!- PEPTIDE MASSES: SPOT 2D-001VN0: 883.4; 932.5; 984.5; 1054.5;
2D 1125.6; 1182.6; 1278.6; 1294.6; 1366.7; 1391.7; 1407.7; 1629.8; 1651.8;
2D 1816.9; 1832.8; 1927; 2185.1; 2348.2; 2476.3; 3235.8; TRYPSIN.
2D -!- MAPPING: Peptide mass fingerprinting [1].
CC ---------------------------------------------------------------------------
CC This SWISS-2DPAGE entry is copyright the Swiss Institute of Bioinformatics.
CC There are no restrictions on its use by non-profit institutions as long as
CC its content is in no way modified and this statement is not removed.
CC Usage by and for commercial entities requires a license agreement (See
CC http://world-2dpage.expasy.org/swiss-2dpage/docs/license.html or send email from legal@sib.swiss).
CC ---------------------------------------------------------------------------
DR UniProtKB/Swiss-Prot; Q9UQ80; PA2G4_HUMAN.
//
The following 2-D maps are available for this protein:
|
SDS-PAGE of nucleolar proteins from Human HeLa cells
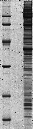 |
2D-PAGE of nucleolar proteins from Human HeLa cells
 |
|---|
SWISS-2DPAGE (search AC)
|