Attention: World-2DPAGE is no longer maintained.
[Click to access protein entry]
Spot: 2D-000KZS (ecoli)
pI: 5.00 Mw: 41546
%vol: 0.15775 %od: 0.253402
================
*PGK_ECOLI*
accession n°: P0A799
Identification Methods:
* MAPPING: {Mi,Aa}
Spot: 2D-000KZS (ecoli)
pI: 5.00 Mw: 41546
%vol: 0.15775 %od: 0.253402
================
*PGK_ECOLI*
accession n°: P0A799
Identification Methods:
* MAPPING: {Mi,Aa}
[Click to access protein entry]
Spot: 2D-000KZ9 (ecoli)
pI: 5.07 Mw: 42043
%vol: 0.177652 %od: 0.520675
================
*PGK_ECOLI*
accession n°: P0A799
Identification Methods:
* MAPPING: {Mi,Gm}
Spot: 2D-000KZ9 (ecoli)
pI: 5.07 Mw: 42043
%vol: 0.177652 %od: 0.520675
================
*PGK_ECOLI*
accession n°: P0A799
Identification Methods:
* MAPPING: {Mi,Gm}
[Click to access protein entry]
Spot: 2D-000KZD (ecoli)
pI: 5.02 Mw: 41877
%vol: 0.127854 %od: 0.248003
================
*PGK_ECOLI*
accession n°: P0A799
Identification Methods:
* MAPPING: {Mi,Aa}
amino acid composition (in %): {
$m/Z=B=9,Z=8.5,S=5.7,H=1.1,G=9.3,T=5.6,A=11.4,P=4.5,Y=2.5,R=4.5,
V=8.8,M=1.6,I=6.1,L=10.1,F=4.4,K=6.9; }
Spot: 2D-000KZD (ecoli)
pI: 5.02 Mw: 41877
%vol: 0.127854 %od: 0.248003
================
*PGK_ECOLI*
accession n°: P0A799
Identification Methods:
* MAPPING: {Mi,Aa}
amino acid composition (in %): {
$m/Z=B=9,Z=8.5,S=5.7,H=1.1,G=9.3,T=5.6,A=11.4,P=4.5,Y=2.5,R=4.5,
V=8.8,M=1.6,I=6.1,L=10.1,F=4.4,K=6.9; }
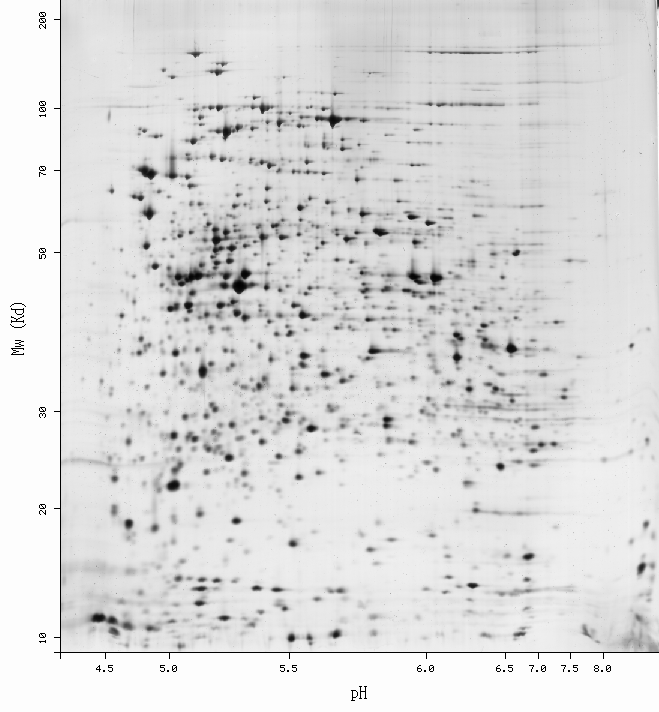
SWISS-2DPAGE Viewer
|
Back to the |
-Get more information by dragging your mouse pointer over any highlighted spot -Click on a highlighted spot to access all its associated protein entries
[estimated location]
Back to the