Attention: World-2DPAGE is no longer maintained.
[Click to access protein entry]
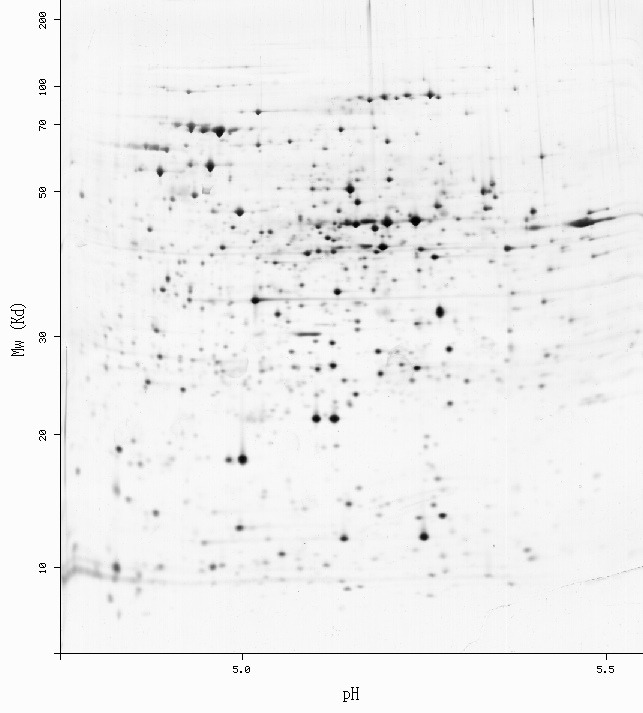
Spot: 2D-0014UQ (ecoli4.5-5.5)
pI: 4.99 Mw: 55445
%vol: 0.493174 %od: 1.41845
================
*CH60_ECOLI*
accession n°: P0A6F5
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 921.46 (0), 985.49 (0), 1011.48 (0), 1201.53 (0), 1215.61 (0),
1249.53 (0), 1454.55 (0), 1567.77 (0), 1589.74 (0), 1782.69 (0),
1845.77 (0), 2042.79 (0), 2402.01 (0), 2805.51 (0), 2867.05 (0)
}
Spot: 2D-0014UQ (ecoli4.5-5.5)
pI: 4.99 Mw: 55445
%vol: 0.493174 %od: 1.41845
================
*CH60_ECOLI*
accession n°: P0A6F5
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 921.46 (0), 985.49 (0), 1011.48 (0), 1201.53 (0), 1215.61 (0),
1249.53 (0), 1454.55 (0), 1567.77 (0), 1589.74 (0), 1782.69 (0),
1845.77 (0), 2042.79 (0), 2402.01 (0), 2805.51 (0), 2867.05 (0)
}
[Click to access protein entry]
Spot: 2D-0014V3 (ecoli4.5-5.5)
pI: 4.96 Mw: 54287
%vol: 0.43502 %od: 0.862776
================
*CH60_ECOLI*
accession n°: P0A6F5
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1011.54 (0), 1201.6 (0), 1454.7 (0), 1567.9 (0), 1846 (0),
2402.3 (0), 2867.4 (0) }
Spot: 2D-0014V3 (ecoli4.5-5.5)
pI: 4.96 Mw: 54287
%vol: 0.43502 %od: 0.862776
================
*CH60_ECOLI*
accession n°: P0A6F5
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1011.54 (0), 1201.6 (0), 1454.7 (0), 1567.9 (0), 1846 (0),
2402.3 (0), 2867.4 (0) }
SWISS-2DPAGE Viewer
|
Back to the |
-Get more information by dragging your mouse pointer over any highlighted spot -Click on a highlighted spot to access all its associated protein entries
[estimated location]
Back to the