Attention: World-2DPAGE is no longer maintained.
[Click to access protein entry]
Spot: 2D-001L27 (ecoli5-6)
pI: 5.23 Mw: 50990
%vol: 0.218986 %od: 0.266145
================
*AROA_ECOLI*
accession n°: P0A6D3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1067.376 (0), 1092.374 (0), 1179.437 (0), 1274.475 (0),
1375.566 (0), 1386.476 (0), 1460.544 (0), 1473.488 (0), 1552.547 (0),
1563.521 (0), 1601.556 (0), 1635.604 (0), 1791.459 (0)
}
Spot: 2D-001L27 (ecoli5-6)
pI: 5.23 Mw: 50990
%vol: 0.218986 %od: 0.266145
================
*AROA_ECOLI*
accession n°: P0A6D3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1067.376 (0), 1092.374 (0), 1179.437 (0), 1274.475 (0),
1375.566 (0), 1386.476 (0), 1460.544 (0), 1473.488 (0), 1552.547 (0),
1563.521 (0), 1601.556 (0), 1635.604 (0), 1791.459 (0)
}
[Click to access protein entry]
Spot: 2D-001KYP (ecoli5-6)
pI: 5.34 Mw: 53476
%vol: 0.102032 %od: 0.103901
================
*AROA_ECOLI*
accession n°: P0A6D3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 865.277 (0), 1067.311 (0), 1274.429 (0), 1375.483 (0),
1386.382 (0), 1460.462 (0), 1473.363 (0), 1563.427 (0), 1601.453 (0),
1635.484 (0), 1791.359 (0) }
Spot: 2D-001KYP (ecoli5-6)
pI: 5.34 Mw: 53476
%vol: 0.102032 %od: 0.103901
================
*AROA_ECOLI*
accession n°: P0A6D3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 865.277 (0), 1067.311 (0), 1274.429 (0), 1375.483 (0),
1386.382 (0), 1460.462 (0), 1473.363 (0), 1563.427 (0), 1601.453 (0),
1635.484 (0), 1791.359 (0) }
SWISS-2DPAGE Viewer
|
Back to the |
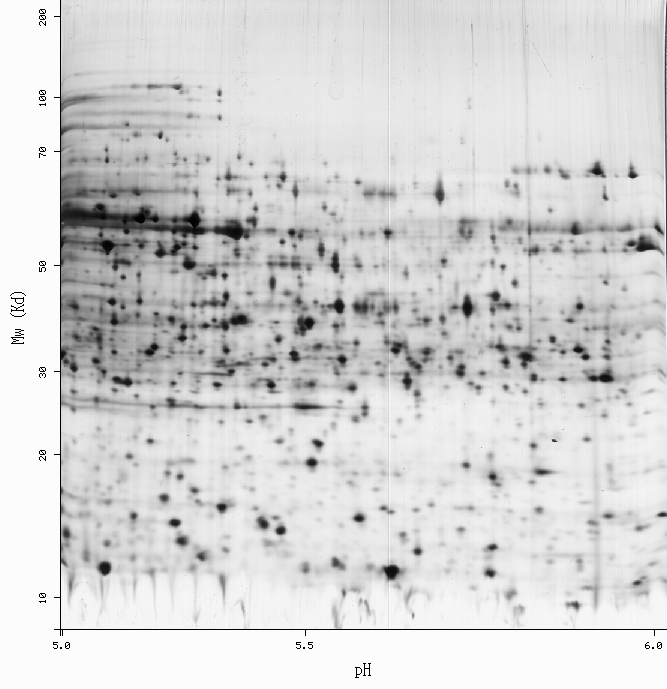
-Get more information by dragging your mouse pointer over any highlighted spot -Click on a highlighted spot to access all its associated protein entries
[estimated location]
Back to the