Attention: World-2DPAGE is no longer maintained.
[Click to access protein entry]
Spot: 2D-001LC7 (ecoli5-6)
pI: 5.51 Mw: 37702
%vol: 0.241842 %od: 0.703903
================
*ZNUA_ECOLI*
accession n°: P39172
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1072.688 (0), 1088.675 (0), 1259.768 (0), 1590.779 (0),
1742.927 (0), 1826.185 (0), 1842.178 (0), 2234.329 (0), 2261.296 (0),
2407.48 (0), 2643.451 (0), 2659.428 (0) }
Spot: 2D-001LC7 (ecoli5-6)
pI: 5.51 Mw: 37702
%vol: 0.241842 %od: 0.703903
================
*ZNUA_ECOLI*
accession n°: P39172
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1072.688 (0), 1088.675 (0), 1259.768 (0), 1590.779 (0),
1742.927 (0), 1826.185 (0), 1842.178 (0), 2234.329 (0), 2261.296 (0),
2407.48 (0), 2643.451 (0), 2659.428 (0) }
[Click to access protein entry]
Spot: 2D-001LFK (ecoli5-6)
pI: 5.03 Mw: 33559
%vol: 0.0748641 %od: 0.0700685
================
*TRPB_ECOLI*
accession n°: P0A879
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 914.558 (0), 930.554 (0), 948.508 (0), 1200.739 (0),
1227.787 (0), 1500.934 (0), 1710.063 (0), 1726.064 (0), 1787.018 (0),
1811.125 (0), 2424.493 (0) }
Spot: 2D-001LFK (ecoli5-6)
pI: 5.03 Mw: 33559
%vol: 0.0748641 %od: 0.0700685
================
*TRPB_ECOLI*
accession n°: P0A879
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 914.558 (0), 930.554 (0), 948.508 (0), 1200.739 (0),
1227.787 (0), 1500.934 (0), 1710.063 (0), 1726.064 (0), 1787.018 (0),
1811.125 (0), 2424.493 (0) }
[Click to access protein entry]
Spot: 2D-001LRH (ecoli5-6)
pI: 5.21 Mw: 16164
%vol: 0.133168 %od: 0.246888
================
*XGPT_ECOLI*
accession n°: P0A9M5
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1023.611 (0), 1034.5 (0), 1888.775 (0), 1945.778 (0),
2490.135 (0), 2646.249 (0) }
================
*DPS_ECOLI*
accession n: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 951.14 (0), 1020.12 (0), 1501.09 (0), 1517.204 (0),
1678.142 (0), 1692.113 (0) }
Spot: 2D-001LRH (ecoli5-6)
pI: 5.21 Mw: 16164
%vol: 0.133168 %od: 0.246888
================
*XGPT_ECOLI*
accession n°: P0A9M5
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1023.611 (0), 1034.5 (0), 1888.775 (0), 1945.778 (0),
2490.135 (0), 2646.249 (0) }
================
*DPS_ECOLI*
accession n: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 951.14 (0), 1020.12 (0), 1501.09 (0), 1517.204 (0),
1678.142 (0), 1692.113 (0) }
[Click to access protein entry]
Spot: 2D-001L4V (ecoli5-6)
pI: 5.53 Mw: 50441
%vol: 0.157146 %od: 0.19062
================
*AAT_ECOLI*
accession n°: P00509
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 869.604 (0), 976.727 (0), 1012.644 (0), 1088.695 (0),
1129.766 (0), 1178.799 (0), 1219.826 (0), 1284.817 (0), 1391.912 (0),
1407.88 (0), 1450.966 (0), 1554.037 (0), 1567.028 (0), 1607.033 (0),
1664.052 (0), 2438.584 (0) }
Spot: 2D-001L4V (ecoli5-6)
pI: 5.53 Mw: 50441
%vol: 0.157146 %od: 0.19062
================
*AAT_ECOLI*
accession n°: P00509
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 869.604 (0), 976.727 (0), 1012.644 (0), 1088.695 (0),
1129.766 (0), 1178.799 (0), 1219.826 (0), 1284.817 (0), 1391.912 (0),
1407.88 (0), 1450.966 (0), 1554.037 (0), 1567.028 (0), 1607.033 (0),
1664.052 (0), 2438.584 (0) }
[Click to access protein entry]
Spot: 2D-001KYY (ecoli5-6)
pI: 5.38 Mw: 52838
%vol: 0.0895264 %od: 0.0748468
================
*SAD_ECOLI*
accession n°: P76149
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 910.394 (0), 1073.57 (0), 1159.51 (0), 1202.551 (0),
1218.53 (0), 1582.715 (0), 1665.744 (0), 1774.724 (0)
}
Spot: 2D-001KYY (ecoli5-6)
pI: 5.38 Mw: 52838
%vol: 0.0895264 %od: 0.0748468
================
*SAD_ECOLI*
accession n°: P76149
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 910.394 (0), 1073.57 (0), 1159.51 (0), 1202.551 (0),
1218.53 (0), 1582.715 (0), 1665.744 (0), 1774.724 (0)
}
[Click to access protein entry]
Spot: 2D-001LKS (ecoli5-6)
pI: 5.76 Mw: 27614
%vol: 0.115401 %od: 0.0841291
================
*GST_ECOLI*
accession n°: P0A9D2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1197.686 (0), 1267.707 (0), 1283.66 (0), 1529.828 (0),
1637.785 (0), 1757.877 (0), 1859.915 (0), 2250.067 (0)
}
Spot: 2D-001LKS (ecoli5-6)
pI: 5.76 Mw: 27614
%vol: 0.115401 %od: 0.0841291
================
*GST_ECOLI*
accession n°: P0A9D2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1197.686 (0), 1267.707 (0), 1283.66 (0), 1529.828 (0),
1637.785 (0), 1757.877 (0), 1859.915 (0), 2250.067 (0)
}
[Click to access protein entry]
Spot: 2D-001LUV (ecoli5-6)
pI: 5.46 Mw: 12295
%vol: 0.0264784 %od: 0.04102
================
*FETP_ECOLI*
accession n°: P0A8P3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 866.381 (0), 1022.456 (0), 1128.495 (0), 1183.466 (0),
1185.524 (0), 1243.497 (0), 1259.508 (0), 1275.497 (0), 1416.577 (0),
1544.62 (0), 1780.683 (0) }
Spot: 2D-001LUV (ecoli5-6)
pI: 5.46 Mw: 12295
%vol: 0.0264784 %od: 0.04102
================
*FETP_ECOLI*
accession n°: P0A8P3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 866.381 (0), 1022.456 (0), 1128.495 (0), 1183.466 (0),
1185.524 (0), 1243.497 (0), 1259.508 (0), 1275.497 (0), 1416.577 (0),
1544.62 (0), 1780.683 (0) }
[Click to access protein entry]
Spot: 2D-001LCD (ecoli5-6)
pI: 5.11 Mw: 37922
%vol: 0.222436 %od: 0.305655
================
*DAPB_ECOLI*
accession n°: P04036
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 954.617 (0), 970.615 (0), 1117.853 (0), 1889.238 (0),
1899.263 (0), 1905.204 (0), 1915.228 (0), 2064.532 (0)
}
Spot: 2D-001LCD (ecoli5-6)
pI: 5.11 Mw: 37922
%vol: 0.222436 %od: 0.305655
================
*DAPB_ECOLI*
accession n°: P04036
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 954.617 (0), 970.615 (0), 1117.853 (0), 1889.238 (0),
1899.263 (0), 1905.204 (0), 1915.228 (0), 2064.532 (0)
}
[Click to access protein entry]
Spot: 2D-001L6G (ecoli5-6)
pI: 5.26 Mw: 50000
%vol: 0.221228 %od: 0.700966
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1189.623 (0), 1255.672 (0), 1459.807 (0), 1562.835 (0),
1605.872 (0), 1826.898 (0), 1928.9 (0), 2856.274 (0)
}
================
*SERC_ECOLI*
accession n: P23721
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 872.508 (0), 945.526 (0), 1002.552 (0), 1069.619 (0),
1162.673 (0), 1214.63 (0), 1285.66 (0), 1322.901 (0), 1342.756 (0),
1357.742 (0), 1373.723 (0), 1392.789 (0), 1408.783 (0), 1442.868 (0),
1548.855 (0), 1564.853 (0), 1602.904 (0), 1645.971 (0), 1710.938 (0),
1774.979 (0), 1802.962 (0), 2020.133 (0), 2036.137 (0)
}
Spot: 2D-001L6G (ecoli5-6)
pI: 5.26 Mw: 50000
%vol: 0.221228 %od: 0.700966
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1189.623 (0), 1255.672 (0), 1459.807 (0), 1562.835 (0),
1605.872 (0), 1826.898 (0), 1928.9 (0), 2856.274 (0)
}
================
*SERC_ECOLI*
accession n: P23721
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 872.508 (0), 945.526 (0), 1002.552 (0), 1069.619 (0),
1162.673 (0), 1214.63 (0), 1285.66 (0), 1322.901 (0), 1342.756 (0),
1357.742 (0), 1373.723 (0), 1392.789 (0), 1408.783 (0), 1442.868 (0),
1548.855 (0), 1564.853 (0), 1602.904 (0), 1645.971 (0), 1710.938 (0),
1774.979 (0), 1802.962 (0), 2020.133 (0), 2036.137 (0)
}
[Click to access protein entry]
Spot: 2D-001L8E (ecoli5-6)
pI: 5.81 Mw: 45820
%vol: 0.114366 %od: 0.124039
================
*SYFA_ECOLI*
accession n°: P08312
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 823.451 (0), 958.517 (0), 989.555 (0), 1027.506 (0),
1156.618 (0), 1272.692 (0), 1310.654 (0), 1341.728 (0), 1600.819 (0),
1890.936 (0), 2320.031 (0) }
Spot: 2D-001L8E (ecoli5-6)
pI: 5.81 Mw: 45820
%vol: 0.114366 %od: 0.124039
================
*SYFA_ECOLI*
accession n°: P08312
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 823.451 (0), 958.517 (0), 989.555 (0), 1027.506 (0),
1156.618 (0), 1272.692 (0), 1310.654 (0), 1341.728 (0), 1600.819 (0),
1890.936 (0), 2320.031 (0) }
[Click to access protein entry]
Spot: 2D-001LNA (ecoli5-6)
pI: 5.33 Mw: 23392
%vol: 0.0578729 %od: 0.0500877
================
*LIVJ_ECOLI*
accession n°: P0AD96
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 908.628 (0), 1639.008 (0), 1767.13 (0), 1785.173 (0),
2401.435 (0), 2417.411 (0) }
Spot: 2D-001LNA (ecoli5-6)
pI: 5.33 Mw: 23392
%vol: 0.0578729 %od: 0.0500877
================
*LIVJ_ECOLI*
accession n°: P0AD96
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 908.628 (0), 1639.008 (0), 1767.13 (0), 1785.173 (0),
2401.435 (0), 2417.411 (0) }
[Click to access protein entry]
Spot: 2D-001LAE (ecoli5-6)
pI: 5.74 Mw: 40430
%vol: 0.236581 %od: 0.940718
================
*SYFA_ECOLI*
accession n°: P08312
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 823.408 (0), 958.481 (0), 989.518 (0), 1027.49 (0),
1156.593 (0), 1192.651 (0), 1310.64 (0), 1341.73 (0), 1600.814 (0),
1890.979 (0) }
================
*G3P1_ECOLI*
accession n: P0A9B2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 835.357 (0), 894.435 (0), 923.465 (0), 1108.633 (0),
1161.611 (0), 1213.652 (0), 1401.733 (0), 1495.839 (0)
}
Spot: 2D-001LAE (ecoli5-6)
pI: 5.74 Mw: 40430
%vol: 0.236581 %od: 0.940718
================
*SYFA_ECOLI*
accession n°: P08312
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 823.408 (0), 958.481 (0), 989.518 (0), 1027.49 (0),
1156.593 (0), 1192.651 (0), 1310.64 (0), 1341.73 (0), 1600.814 (0),
1890.979 (0) }
================
*G3P1_ECOLI*
accession n: P0A9B2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 835.357 (0), 894.435 (0), 923.465 (0), 1108.633 (0),
1161.611 (0), 1213.652 (0), 1401.733 (0), 1495.839 (0)
}
[Click to access protein entry]
Spot: 2D-001LKH (ecoli5-6)
pI: 5.32 Mw: 28018
%vol: 0.181209 %od: 0.214983
================
*NFNB_ECOLI*
accession n°: P38489
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 923.425 (0), 939.403 (0), 1051.53 (0), 1067.51 (0),
1094.537 (0), 1156.626 (0), 1216.592 (0), 1227.59 (0), 1284.718 (0),
1330.603 (0), 1346.59 (0), 1370.783 (0), 1458.661 (0), 1492.773 (0),
2566.285 (0) }
Spot: 2D-001LKH (ecoli5-6)
pI: 5.32 Mw: 28018
%vol: 0.181209 %od: 0.214983
================
*NFNB_ECOLI*
accession n°: P38489
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 923.425 (0), 939.403 (0), 1051.53 (0), 1067.51 (0),
1094.537 (0), 1156.626 (0), 1216.592 (0), 1227.59 (0), 1284.718 (0),
1330.603 (0), 1346.59 (0), 1370.783 (0), 1458.661 (0), 1492.773 (0),
2566.285 (0) }
[Click to access protein entry]
Spot: 2D-001LR4 (ecoli5-6)
pI: 5.27 Mw: 16787
%vol: 0.11773 %od: 0.137014
================
*DPS_ECOLI*
accession n°: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 915.56 (0), 951.536 (0), 1020.57 (0), 1043.61 (0), 1501.652 (0),
1517.949 (0), 1678.831 (0), 1692.796 (0) }
Spot: 2D-001LR4 (ecoli5-6)
pI: 5.27 Mw: 16787
%vol: 0.11773 %od: 0.137014
================
*DPS_ECOLI*
accession n°: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 915.56 (0), 951.536 (0), 1020.57 (0), 1043.61 (0), 1501.652 (0),
1517.949 (0), 1678.831 (0), 1692.796 (0) }
[Click to access protein entry]
Spot: 2D-001LOE (ecoli5-6)
pI: 5.18 Mw: 21250
%vol: 0.129977 %od: 0.113239
================
*AROK_ECOLI*
accession n°: P0A6D7
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 945.483 (0), 1143.611 (0), 1314.784 (0), 1350.775 (0),
1442.905 (0), 1483.769 (0), 1532.855 (0), 2485.251 (0)
}
Spot: 2D-001LOE (ecoli5-6)
pI: 5.18 Mw: 21250
%vol: 0.129977 %od: 0.113239
================
*AROK_ECOLI*
accession n°: P0A6D7
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 945.483 (0), 1143.611 (0), 1314.784 (0), 1350.775 (0),
1442.905 (0), 1483.769 (0), 1532.855 (0), 2485.251 (0)
}
[Click to access protein entry]
Spot: 2D-001L61 (ecoli5-6)
pI: 5.00 Mw: 50085
%vol: 0.153609 %od: 0.066609
================
*MALE_ECOLI*
accession n°: P0AEX9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 959.495 (0), 1057.542 (0), 1189.69 (0), 1267.635 (0),
1336.618 (0), 1337.676 (0), 1423.719 (0), 1766.866 (0), 1891.056 (0),
1897.809 (0), 2097.038 (0), 2110.063 (0), 2213.082 (0)
}
Spot: 2D-001L61 (ecoli5-6)
pI: 5.00 Mw: 50085
%vol: 0.153609 %od: 0.066609
================
*MALE_ECOLI*
accession n°: P0AEX9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 959.495 (0), 1057.542 (0), 1189.69 (0), 1267.635 (0),
1336.618 (0), 1337.676 (0), 1423.719 (0), 1766.866 (0), 1891.056 (0),
1897.809 (0), 2097.038 (0), 2110.063 (0), 2213.082 (0)
}
[Click to access protein entry]
Spot: 2D-001L2C (ecoli5-6)
pI: 5.13 Mw: 50973
%vol: 0.219503 %od: 0.226745
================
*OPPA_ECOLI*
accession n°: P23843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 917.63 (0), 958.722 (0), 1080.711 (0), 1170.823 (0),
1200.841 (0), 1274.979 (0), 1425.929 (0), 1636.052 (0), 1764.174 (0),
2143.352 (0), 2232.603 (0), 2337.54 (0), 2606.706 (0)
}
Spot: 2D-001L2C (ecoli5-6)
pI: 5.13 Mw: 50973
%vol: 0.219503 %od: 0.226745
================
*OPPA_ECOLI*
accession n°: P23843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 917.63 (0), 958.722 (0), 1080.711 (0), 1170.823 (0),
1200.841 (0), 1274.979 (0), 1425.929 (0), 1636.052 (0), 1764.174 (0),
2143.352 (0), 2232.603 (0), 2337.54 (0), 2606.706 (0)
}
[Click to access protein entry]
Spot: 2D-001L9U (ecoli5-6)
pI: 5.10 Mw: 41867
%vol: 0.215536 %od: 0.3419
================
*CYSK_ECOLI*
accession n°: P0ABK5
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1186.634 (0), 1210.589 (0), 1247.641 (0), 1256.749 (0),
1263.691 (0), 1283.787 (0), 1403.776 (0), 1473.851 (0), 1521.834 (0),
1537.798 (0), 1553.816 (0), 1618.9 (0), 1626.976 (0), 2141.155 (0),
2257.293 (0), 2273.278 (0), 2753.565 (0), 3367.848 (0)
}
Spot: 2D-001L9U (ecoli5-6)
pI: 5.10 Mw: 41867
%vol: 0.215536 %od: 0.3419
================
*CYSK_ECOLI*
accession n°: P0ABK5
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1186.634 (0), 1210.589 (0), 1247.641 (0), 1256.749 (0),
1263.691 (0), 1283.787 (0), 1403.776 (0), 1473.851 (0), 1521.834 (0),
1537.798 (0), 1553.816 (0), 1618.9 (0), 1626.976 (0), 2141.155 (0),
2257.293 (0), 2273.278 (0), 2753.565 (0), 3367.848 (0)
}
[Click to access protein entry]
Spot: 2D-001LBR (ecoli5-6)
pI: 5.71 Mw: 38478
%vol: 0.11704 %od: 0.158632
================
*AMPM_ECOLI*
accession n°: P0AE18
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1048.494 (0), 1118.555 (0), 1239.687 (0), 1306.785 (0),
1478.822 (0), 1713.841 (0), 2113.063 (0), 2619.318 (0)
}
Spot: 2D-001LBR (ecoli5-6)
pI: 5.71 Mw: 38478
%vol: 0.11704 %od: 0.158632
================
*AMPM_ECOLI*
accession n°: P0AE18
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1048.494 (0), 1118.555 (0), 1239.687 (0), 1306.785 (0),
1478.822 (0), 1713.841 (0), 2113.063 (0), 2619.318 (0)
}
[Click to access protein entry]
Spot: 2D-001LHP (ecoli5-6)
pI: 5.80 Mw: 31295
%vol: 0.187419 %od: 0.221202
================
*CMOA_ECOLI*
accession n°: P76290
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1037.411 (0), 1090.561 (0) }
Spot: 2D-001LHP (ecoli5-6)
pI: 5.80 Mw: 31295
%vol: 0.187419 %od: 0.221202
================
*CMOA_ECOLI*
accession n°: P76290
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1037.411 (0), 1090.561 (0) }
[Click to access protein entry]
Spot: 2D-001LC6 (ecoli5-6)
pI: 5.49 Mw: 38144
%vol: 0.159215 %od: 0.191573
================
*IMDH_ECOLI*
accession n°: P0ADG7
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1217.688 (0), 1257.669 (0), 1302.672 (0), 1786.983 (0),
1855.026 (0), 1986.145 (0), 2179.223 (0), 2335.299 (0), 2757.502 (0)
}
================
*MDH_ECOLI*
accession n: P61889
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 899.183 (0), 1008.15 (0), 1149.214 (0), 1193.177 (0),
1234.166 (0), 1247.292 (0), 1276.205 (0), 1414.244 (0), 1421.138 (0),
1734.384 (0) }
Spot: 2D-001LC6 (ecoli5-6)
pI: 5.49 Mw: 38144
%vol: 0.159215 %od: 0.191573
================
*IMDH_ECOLI*
accession n°: P0ADG7
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1217.688 (0), 1257.669 (0), 1302.672 (0), 1786.983 (0),
1855.026 (0), 1986.145 (0), 2179.223 (0), 2335.299 (0), 2757.502 (0)
}
================
*MDH_ECOLI*
accession n: P61889
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 899.183 (0), 1008.15 (0), 1149.214 (0), 1193.177 (0),
1234.166 (0), 1247.292 (0), 1276.205 (0), 1414.244 (0), 1421.138 (0),
1734.384 (0) }
[Click to access protein entry]
Spot: 2D-001L4G (ecoli5-6)
pI: 5.23 Mw: 50424
%vol: 0.236581 %od: 0.52785
================
*EFG_ECOLI*
accession n°: P0A6M8
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 902.487 (0), 1045.588 (0), 1089.561 (0), 1098.579 (0),
1112.585 (0), 1254.745 (0), 1414.815 (0), 1515.862 (0), 1531.868 (0),
1732.929 (0), 1820.996 (0), 1951.997 (0), 2059.074 (0), 2062.139 (0),
2078.124 (0), 2101.098 (0), 2201.174 (0), 2217.135 (0)
}
================
*LIVJ_ECOLI*
accession n: P0AD96
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 862.52 (0), 908.484 (0), 1021.503 (0), 1361.814 (0),
1638.857 (0), 1784.885 (0) }
Spot: 2D-001L4G (ecoli5-6)
pI: 5.23 Mw: 50424
%vol: 0.236581 %od: 0.52785
================
*EFG_ECOLI*
accession n°: P0A6M8
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 902.487 (0), 1045.588 (0), 1089.561 (0), 1098.579 (0),
1112.585 (0), 1254.745 (0), 1414.815 (0), 1515.862 (0), 1531.868 (0),
1732.929 (0), 1820.996 (0), 1951.997 (0), 2059.074 (0), 2062.139 (0),
2078.124 (0), 2101.098 (0), 2201.174 (0), 2217.135 (0)
}
================
*LIVJ_ECOLI*
accession n: P0AD96
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 862.52 (0), 908.484 (0), 1021.503 (0), 1361.814 (0),
1638.857 (0), 1784.885 (0) }
[Click to access protein entry]
Spot: 2D-001KVT (ecoli5-6)
pI: 5.00 Mw: 74518
%vol: 0.0615817 %od: 0.0211558
================
*IDH_ECOLI*
accession n°: P08200
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1008.45 (0), 1030.433 (0), 1124.616 (0), 1206.587 (0),
1357.692 (0), 1470.752 (0), 1484.719 (0), 1553.721 (0), 1630.906 (0),
1646.851 (0), 1989.021 (0), 2086.181 (0), 2202.183 (0), 2304.309 (0)
}
Spot: 2D-001KVT (ecoli5-6)
pI: 5.00 Mw: 74518
%vol: 0.0615817 %od: 0.0211558
================
*IDH_ECOLI*
accession n°: P08200
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1008.45 (0), 1030.433 (0), 1124.616 (0), 1206.587 (0),
1357.692 (0), 1470.752 (0), 1484.719 (0), 1553.721 (0), 1630.906 (0),
1646.851 (0), 1989.021 (0), 2086.181 (0), 2202.183 (0), 2304.309 (0)
}
[Click to access protein entry]
Spot: 2D-001L2S (ecoli5-6)
pI: 5.47 Mw: 50921
%vol: 0.201995 %od: 0.243344
================
*OPPA_ECOLI*
accession n°: P23843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 917.462 (0), 958.549 (0), 1059.619 (0), 1080.525 (0),
1187.698 (0), 1200.659 (0), 1273.704 (0), 1635.805 (0), 1763.894 (0),
1791.892 (0), 2001.916 (0), 2053.939 (0), 2143.011 (0), 2232.213 (0),
2337.177 (0), 2606.315 (0), 2781.321 (0), 2874.477 (0), 3727.719 (0)
}
Spot: 2D-001L2S (ecoli5-6)
pI: 5.47 Mw: 50921
%vol: 0.201995 %od: 0.243344
================
*OPPA_ECOLI*
accession n°: P23843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 917.462 (0), 958.549 (0), 1059.619 (0), 1080.525 (0),
1187.698 (0), 1200.659 (0), 1273.704 (0), 1635.805 (0), 1763.894 (0),
1791.892 (0), 2001.916 (0), 2053.939 (0), 2143.011 (0), 2232.213 (0),
2337.177 (0), 2606.315 (0), 2781.321 (0), 2874.477 (0), 3727.719 (0)
}
[Click to access protein entry]
Spot: 2D-001L06 (ecoli5-6)
pI: 5.29 Mw: 51580
%vol: 0.0683955 %od: 0.0519998
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 950.606 (0), 1189.757 (0), 1255.782 (0), 1459.977 (0),
1563.017 (0), 1929.113 (0) }
Spot: 2D-001L06 (ecoli5-6)
pI: 5.29 Mw: 51580
%vol: 0.0683955 %od: 0.0519998
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 950.606 (0), 1189.757 (0), 1255.782 (0), 1459.977 (0),
1563.017 (0), 1929.113 (0) }
[Click to access protein entry]
Spot: 2D-001LU0 (ecoli5-6)
pI: 5.25 Mw: 13108
%vol: 0.161027 %od: 0.367934
================
*OMPA_ECOLI*
accession n°: P0A910
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 872.527 (0), 1083.557 (0), 1222.675 (0), 1280.675 (0),
1378.789 (0), 1409.693 (0) }
Spot: 2D-001LU0 (ecoli5-6)
pI: 5.25 Mw: 13108
%vol: 0.161027 %od: 0.367934
================
*OMPA_ECOLI*
accession n°: P0A910
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 872.527 (0), 1083.557 (0), 1222.675 (0), 1280.675 (0),
1378.789 (0), 1409.693 (0) }
[Click to access protein entry]
Spot: 2D-001LEF (ecoli5-6)
pI: 5.25 Mw: 35056
%vol: 0.211224 %od: 0.309324
================
*PANC_ECOLI*
accession n°: P31663
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1017.755 (0), 1022.638 (0), 1051.718 (0), 1103.83 (0),
1179.835 (0), 1202.923 (0), 1320.844 (0), 1355.008 (0), 1487.089 (0),
1564.074 (0), 1751.201 (0), 1801.255 (0), 1908.302 (0)
}
Spot: 2D-001LEF (ecoli5-6)
pI: 5.25 Mw: 35056
%vol: 0.211224 %od: 0.309324
================
*PANC_ECOLI*
accession n°: P31663
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1017.755 (0), 1022.638 (0), 1051.718 (0), 1103.83 (0),
1179.835 (0), 1202.923 (0), 1320.844 (0), 1355.008 (0), 1487.089 (0),
1564.074 (0), 1751.201 (0), 1801.255 (0), 1908.302 (0)
}
[Click to access protein entry]
Spot: 2D-001KZT (ecoli5-6)
pI: 5.01 Mw: 51702
%vol: 0.154213 %od: 0.205852
================
*ACEA_ECOLI*
accession n°: P0A9G6
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1041.586 (0), 1088.541 (0), 1182.624 (0), 1225.688 (0),
1495.885 (0), 1716.804 (0), 1732.855 (0), 1739.822 (0), 1749.877 (0),
1895.942 (0), 2018.098 (0), 2058.025 (0), 2177.978 (0), 2711.303 (0),
2777.167 (0) }
Spot: 2D-001KZT (ecoli5-6)
pI: 5.01 Mw: 51702
%vol: 0.154213 %od: 0.205852
================
*ACEA_ECOLI*
accession n°: P0A9G6
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1041.586 (0), 1088.541 (0), 1182.624 (0), 1225.688 (0),
1495.885 (0), 1716.804 (0), 1732.855 (0), 1739.822 (0), 1749.877 (0),
1895.942 (0), 2018.098 (0), 2058.025 (0), 2177.978 (0), 2711.303 (0),
2777.167 (0) }
[Click to access protein entry]
Spot: 2D-001LFC (ecoli5-6)
pI: 5.52 Mw: 33657
%vol: 0.144036 %od: 0.223182
================
*DEOC_ECOLI*
accession n°: P0A6L0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 891.536 (0), 907.514 (0), 923.501 (0), 929.514 (0),
1088.688 (0), 1114.657 (0), 1206.767 (0), 1271.759 (0), 1475.852 (0),
1532.89 (0), 1607.898 (0), 1699.069 (0), 1856.031 (0), 1897.027 (0),
2383.295 (0) }
Spot: 2D-001LFC (ecoli5-6)
pI: 5.52 Mw: 33657
%vol: 0.144036 %od: 0.223182
================
*DEOC_ECOLI*
accession n°: P0A6L0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 891.536 (0), 907.514 (0), 923.501 (0), 929.514 (0),
1088.688 (0), 1114.657 (0), 1206.767 (0), 1271.759 (0), 1475.852 (0),
1532.89 (0), 1607.898 (0), 1699.069 (0), 1856.031 (0), 1897.027 (0),
2383.295 (0) }
[Click to access protein entry]
Spot: 2D-001LHC (ecoli5-6)
pI: 5.48 Mw: 31661
%vol: 0.186901 %od: 0.205752
================
*KAD_ECOLI*
accession n°: P69441
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 879.503 (0), 891.421 (0), 974.56 (0), 990.502 (0), 1009.654 (0),
1015.571 (0), 1119.561 (0), 1135.607 (0), 1152.624 (0), 1168.586 (0),
1170.683 (0), 1235.618 (0), 1312.829 (0), 1450.781 (0), 1466.77 (0),
1648.837 (0), 2013.062 (0), 2029.03 (0), 2520.305 (0)
}
Spot: 2D-001LHC (ecoli5-6)
pI: 5.48 Mw: 31661
%vol: 0.186901 %od: 0.205752
================
*KAD_ECOLI*
accession n°: P69441
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 879.503 (0), 891.421 (0), 974.56 (0), 990.502 (0), 1009.654 (0),
1015.571 (0), 1119.561 (0), 1135.607 (0), 1152.624 (0), 1168.586 (0),
1170.683 (0), 1235.618 (0), 1312.829 (0), 1450.781 (0), 1466.77 (0),
1648.837 (0), 2013.062 (0), 2029.03 (0), 2520.305 (0)
}
[Click to access protein entry]
Spot: 2D-001L2T (ecoli5-6)
pI: 5.63 Mw: 50921
%vol: 0.188195 %od: 0.228756
================
*OPPA_ECOLI*
accession n°: P23843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 917.559 (0), 958.649 (0), 1059.728 (0), 1080.647 (0),
1170.77 (0), 1200.774 (0), 1274.905 (0), 1635.959 (0), 1764.101 (0),
2143.244 (0), 2232.448 (0), 2337.422 (0), 2606.568 (0)
}
Spot: 2D-001L2T (ecoli5-6)
pI: 5.63 Mw: 50921
%vol: 0.188195 %od: 0.228756
================
*OPPA_ECOLI*
accession n°: P23843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 917.559 (0), 958.649 (0), 1059.728 (0), 1080.647 (0),
1170.77 (0), 1200.774 (0), 1274.905 (0), 1635.959 (0), 1764.101 (0),
2143.244 (0), 2232.448 (0), 2337.422 (0), 2606.568 (0)
}
[Click to access protein entry]
Spot: 2D-001LDG (ecoli5-6)
pI: 5.81 Mw: 36727
%vol: 0.112124 %od: 0.0785857
================
*ATPA_ECOLI*
accession n°: P0ABB0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 882.533 (0), 946.554 (0), 1062.608 (0), 1074.568 (0),
1298.724 (0), 1537.692 (0), 1722.776 (0), 2149.925 (0), 2641.087 (0)
}
Spot: 2D-001LDG (ecoli5-6)
pI: 5.81 Mw: 36727
%vol: 0.112124 %od: 0.0785857
================
*ATPA_ECOLI*
accession n°: P0ABB0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 882.533 (0), 946.554 (0), 1062.608 (0), 1074.568 (0),
1298.724 (0), 1537.692 (0), 1722.776 (0), 2149.925 (0), 2641.087 (0)
}
[Click to access protein entry]
Spot: 2D-001LD3 (ecoli5-6)
pI: 5.25 Mw: 37049
%vol: 0.19958 %od: 0.204167
================
*MDH_ECOLI*
accession n°: P61889
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 899.676 (0), 1008.707 (0), 1149.87 (0), 1193.867 (0),
1247.996 (0), 1276.926 (0), 1405.977 (0), 1415.043 (0), 1421.945 (0),
1735.358 (0), 2399.536 (0), 2402.648 (0), 2643.872 (0), 3376.377 (0)
}
Spot: 2D-001LD3 (ecoli5-6)
pI: 5.25 Mw: 37049
%vol: 0.19958 %od: 0.204167
================
*MDH_ECOLI*
accession n°: P61889
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 899.676 (0), 1008.707 (0), 1149.87 (0), 1193.867 (0),
1247.996 (0), 1276.926 (0), 1405.977 (0), 1415.043 (0), 1421.945 (0),
1735.358 (0), 2399.536 (0), 2402.648 (0), 2643.872 (0), 3376.377 (0)
}
[Click to access protein entry]
Spot: 2D-001LH9 (ecoli5-6)
pI: 5.66 Mw: 31754
%vol: 0.133772 %od: 0.121881
================
*UDP_ECOLI*
accession n°: P12758
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 861.262 (0), 1086.661 (0), 1102.636 (0), 1116.635 (0),
1752.1 (0), 2108.372 (0), 2419.671 (0) }
Spot: 2D-001LH9 (ecoli5-6)
pI: 5.66 Mw: 31754
%vol: 0.133772 %od: 0.121881
================
*UDP_ECOLI*
accession n°: P12758
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 861.262 (0), 1086.661 (0), 1102.636 (0), 1116.635 (0),
1752.1 (0), 2108.372 (0), 2419.671 (0) }
[Click to access protein entry]
Spot: 2D-001L2E (ecoli5-6)
pI: 5.35 Mw: 50921
%vol: 0.253831 %od: 1.61302
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.725 (0), 1130.675 (0), 1171.788 (0), 1203.668 (0),
1233.786 (0), 1376.81 (0), 1490.054 (0), 1575.135 (0), 1689.945 (0),
1769.026 (0), 1781.199 (0), 1796.181 (0), 1804.163 (0), 1813.118 (0),
1962.272 (0), 2117.446 (0) }
Spot: 2D-001L2E (ecoli5-6)
pI: 5.35 Mw: 50921
%vol: 0.253831 %od: 1.61302
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.725 (0), 1130.675 (0), 1171.788 (0), 1203.668 (0),
1233.786 (0), 1376.81 (0), 1490.054 (0), 1575.135 (0), 1689.945 (0),
1769.026 (0), 1781.199 (0), 1796.181 (0), 1804.163 (0), 1813.118 (0),
1962.272 (0), 2117.446 (0) }
[Click to access protein entry]
Spot: 2D-001LKD (ecoli5-6)
pI: 5.43 Mw: 27937
%vol: 0.209671 %od: 0.304658
================
*ALKH_ECOLI*
accession n°: P0A955
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 829.528 (0), 977.441 (0), 1136.548 (0), 1357.761 (0),
1910.097 (0) }
Spot: 2D-001LKD (ecoli5-6)
pI: 5.43 Mw: 27937
%vol: 0.209671 %od: 0.304658
================
*ALKH_ECOLI*
accession n°: P0A955
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 829.528 (0), 977.441 (0), 1136.548 (0), 1357.761 (0),
1910.097 (0) }
[Click to access protein entry]
Spot: 2D-001LOD (ecoli5-6)
pI: 5.45 Mw: 21312
%vol: 0.0803842 %od: 0.0942542
================
*SODF_ECOLI*
accession n°: P0AGD3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1235.561 (0), 1630.805 (0), 1694.876 (0), 2015.019 (0)
}
Spot: 2D-001LOD (ecoli5-6)
pI: 5.45 Mw: 21312
%vol: 0.0803842 %od: 0.0942542
================
*SODF_ECOLI*
accession n°: P0AGD3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1235.561 (0), 1630.805 (0), 1694.876 (0), 2015.019 (0)
}
[Click to access protein entry]
Spot: 2D-001KZX (ecoli5-6)
pI: 5.08 Mw: 51667
%vol: 0.235891 %od: 0.375224
================
*ASSY_ECOLI*
accession n°: P0A6E4
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 910.432 (0), 1013.56 (0), 1094.494 (0), 1111.5 (0),
1127.507 (0), 1266.645 (0), 1282.622 (0), 1356.633 (0), 1372.616 (0),
1540.715 (0), 1673.728 (0), 1701.901 (0), 1903.939 (0), 1919.932 (0),
2101.029 (0), 2204.07 (0), 2313.108 (0), 2625.184 (0)
}
Spot: 2D-001KZX (ecoli5-6)
pI: 5.08 Mw: 51667
%vol: 0.235891 %od: 0.375224
================
*ASSY_ECOLI*
accession n°: P0A6E4
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 910.432 (0), 1013.56 (0), 1094.494 (0), 1111.5 (0),
1127.507 (0), 1266.645 (0), 1282.622 (0), 1356.633 (0), 1372.616 (0),
1540.715 (0), 1673.728 (0), 1701.901 (0), 1903.939 (0), 1919.932 (0),
2101.029 (0), 2204.07 (0), 2313.108 (0), 2625.184 (0)
}
[Click to access protein entry]
Spot: 2D-001LH3 (ecoli5-6)
pI: 5.72 Mw: 31846
%vol: 0.126096 %od: 0.112635
================
*ACKA_ECOLI*
accession n°: P0A6A3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1102.49 (0), 1330.642 (0), 1346.638 (0), 1370.713 (0),
1835.942 (0), 1838.925 (0), 1866.879 (0), 2022.965 (0), 2465.251 (0)
}
Spot: 2D-001LH3 (ecoli5-6)
pI: 5.72 Mw: 31846
%vol: 0.126096 %od: 0.112635
================
*ACKA_ECOLI*
accession n°: P0A6A3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1102.49 (0), 1330.642 (0), 1346.638 (0), 1370.713 (0),
1835.942 (0), 1838.925 (0), 1866.879 (0), 2022.965 (0), 2465.251 (0)
}
[Click to access protein entry]
Spot: 2D-001LKF (ecoli5-6)
pI: 5.75 Mw: 28100
%vol: 0.0898714 %od: 0.0689373
================
*PURK_ECOLI*
accession n°: P09029
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 842.509 (0), 973.501 (0), 1045.564 (0), 1106.522 (0)
}
Spot: 2D-001LKF (ecoli5-6)
pI: 5.75 Mw: 28100
%vol: 0.0898714 %od: 0.0689373
================
*PURK_ECOLI*
accession n°: P09029
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 842.509 (0), 973.501 (0), 1045.564 (0), 1106.522 (0)
}
[Click to access protein entry]
Spot: 2D-001LDV (ecoli5-6)
pI: 5.71 Mw: 35882
%vol: 0.0911651 %od: 0.084982
================
*SUHB_ECOLI*
accession n°: P0ADG4
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1171.664 (0), 1209.781 (0), 1299.963 (0), 1366.047 (0),
1371.026 (0), 1436.121 (0) }
Spot: 2D-001LDV (ecoli5-6)
pI: 5.71 Mw: 35882
%vol: 0.0911651 %od: 0.084982
================
*SUHB_ECOLI*
accession n°: P0ADG4
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1171.664 (0), 1209.781 (0), 1299.963 (0), 1366.047 (0),
1371.026 (0), 1436.121 (0) }
[Click to access protein entry]
Spot: 2D-001LL5 (ecoli5-6)
pI: 5.67 Mw: 26821
%vol: 0.107725 %od: 0.126332
================
*RISA_ECOLI*
accession n°: P0AFU8
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 946.498 (0), 1317.727 (0), 1456.768 (0), 1931.99 (0),
2025.982 (0) }
Spot: 2D-001LL5 (ecoli5-6)
pI: 5.67 Mw: 26821
%vol: 0.107725 %od: 0.126332
================
*RISA_ECOLI*
accession n°: P0AFU8
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 946.498 (0), 1317.727 (0), 1456.768 (0), 1931.99 (0),
2025.982 (0) }
[Click to access protein entry]
Spot: 2D-001LAM (ecoli5-6)
pI: 5.60 Mw: 40548
%vol: 0.158267 %od: 0.250774
================
*YHDH_ECOLI*
accession n°: P26646
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 915.652 (0), 1006.651 (0), 1148.857 (0), 1319.876 (0),
1320.891 (0), 1359.913 (0), 1373.966 (0), 1389.94 (0), 1428.991 (0),
1444.959 (0), 1585.11 (0), 1601.066 (0), 1635.16 (0), 1651.118 (0),
1653.128 (0), 1756.249 (0), 1772.222 (0), 2414.661 (0)
}
Spot: 2D-001LAM (ecoli5-6)
pI: 5.60 Mw: 40548
%vol: 0.158267 %od: 0.250774
================
*YHDH_ECOLI*
accession n°: P26646
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 915.652 (0), 1006.651 (0), 1148.857 (0), 1319.876 (0),
1320.891 (0), 1359.913 (0), 1373.966 (0), 1389.94 (0), 1428.991 (0),
1444.959 (0), 1585.11 (0), 1601.066 (0), 1635.16 (0), 1651.118 (0),
1653.128 (0), 1756.249 (0), 1772.222 (0), 2414.661 (0)
}
[Click to access protein entry]
Spot: 2D-001LP8 (ecoli5-6)
pI: 5.51 Mw: 19192
%vol: 0.196389 %od: 0.404993
================
*PPIB_ECOLI*
accession n°: P23869
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 930.407 (0), 987.435 (0), 1014.455 (0), 1090.528 (0),
1250.609 (0), 1397.653 (0), 1611.825 (0), 1627.815 (0), 2315.068 (0)
}
Spot: 2D-001LP8 (ecoli5-6)
pI: 5.51 Mw: 19192
%vol: 0.196389 %od: 0.404993
================
*PPIB_ECOLI*
accession n°: P23869
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 930.407 (0), 987.435 (0), 1014.455 (0), 1090.528 (0),
1250.609 (0), 1397.653 (0), 1611.825 (0), 1627.815 (0), 2315.068 (0)
}
[Click to access protein entry]
Spot: 2D-001LBQ (ecoli5-6)
pI: 5.37 Mw: 38366
%vol: 0.237012 %od: 0.646143
================
*FABI_ECOLI*
accession n°: P0AEK4
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 997.596 (0), 1295.619 (0), 1311.587 (0), 1327.597 (0),
1430.782 (0), 1446.743 (0), 1485.682 (0), 1655.827 (0), 1993.03 (0),
2009.011 (0), 2797.251 (0) }
Spot: 2D-001LBQ (ecoli5-6)
pI: 5.37 Mw: 38366
%vol: 0.237012 %od: 0.646143
================
*FABI_ECOLI*
accession n°: P0AEK4
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 997.596 (0), 1295.619 (0), 1311.587 (0), 1327.597 (0),
1430.782 (0), 1446.743 (0), 1485.682 (0), 1655.827 (0), 1993.03 (0),
2009.011 (0), 2797.251 (0) }
[Click to access protein entry]
Spot: 2D-001LIF (ecoli5-6)
pI: 5.73 Mw: 30486
%vol: 0.161803 %od: 0.156796
================
*OTC2_ECOLI*
accession n°: P06960
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1045.514 (0), 1061.474 (0), 1216.686 (0), 1994.116 (0)
}
Spot: 2D-001LIF (ecoli5-6)
pI: 5.73 Mw: 30486
%vol: 0.161803 %od: 0.156796
================
*OTC2_ECOLI*
accession n°: P06960
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1045.514 (0), 1061.474 (0), 1216.686 (0), 1994.116 (0)
}
[Click to access protein entry]
Spot: 2D-001L5L (ecoli5-6)
pI: 5.56 Mw: 50220
%vol: 0.18233 %od: 0.218018
================
*ALF_ECOLI*
accession n°: P0AB71
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1320.801 (0), 1502.988 (0), 1878.257 (0), 2591.649 (0),
2719.736 (0), 2871.916 (0) }
Spot: 2D-001L5L (ecoli5-6)
pI: 5.56 Mw: 50220
%vol: 0.18233 %od: 0.218018
================
*ALF_ECOLI*
accession n°: P0AB71
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1320.801 (0), 1502.988 (0), 1878.257 (0), 2591.649 (0),
2719.736 (0), 2871.916 (0) }
[Click to access protein entry]
Spot: 2D-001LOG (ecoli5-6)
pI: 5.52 Mw: 21066
%vol: 0.182158 %od: 0.30651
================
*LOLA_ECOLI*
accession n°: P61316
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 934.696 (0), 1238.854 (0), 1406.991 (0), 1422.986 (0),
1510.988 (0), 1708.166 (0), 1846.199 (0), 1917.307 (0)
}
Spot: 2D-001LOG (ecoli5-6)
pI: 5.52 Mw: 21066
%vol: 0.182158 %od: 0.30651
================
*LOLA_ECOLI*
accession n°: P61316
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 934.696 (0), 1238.854 (0), 1406.991 (0), 1422.986 (0),
1510.988 (0), 1708.166 (0), 1846.199 (0), 1917.307 (0)
}
[Click to access protein entry]
Spot: 2D-001LSX (ecoli5-6)
pI: 5.53 Mw: 14556
%vol: 0.0195785 %od: 0.017515
================
*NDK_ECOLI*
accession n°: P0A763
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1003.529 (0), 1213.615 (0), 1667.942 (0), 1673.801 (0),
1961.121 (0) }
Spot: 2D-001LSX (ecoli5-6)
pI: 5.53 Mw: 14556
%vol: 0.0195785 %od: 0.017515
================
*NDK_ECOLI*
accession n°: P0A763
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1003.529 (0), 1213.615 (0), 1667.942 (0), 1673.801 (0),
1961.121 (0) }
[Click to access protein entry]
Spot: 2D-001L65 (ecoli5-6)
pI: 5.71 Mw: 50118
%vol: 0.12601 %od: 0.118563
================
*DPPA_ECOLI*
accession n°: P23847
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 937.507 (0), 971.484 (0), 1050.58 (0), 1327.769 (0),
1526.752 (0), 1890.906 (0), 2060.12 (0), 2407.161 (0), 2643.448 (0)
}
Spot: 2D-001L65 (ecoli5-6)
pI: 5.71 Mw: 50118
%vol: 0.12601 %od: 0.118563
================
*DPPA_ECOLI*
accession n°: P23847
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 937.507 (0), 971.484 (0), 1050.58 (0), 1327.769 (0),
1526.752 (0), 1890.906 (0), 2060.12 (0), 2407.161 (0), 2643.448 (0)
}
[Click to access protein entry]
Spot: 2D-001LGC (ecoli5-6)
pI: 5.18 Mw: 32691
%vol: 0.241066 %od: 0.36004
================
*TRPA_ECOLI*
accession n°: P0A877
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 874.557 (0), 972.647 (0), 1098.73 (0), 1146.829 (0),
1349.877 (0), 1359.779 (0), 1543.024 (0), 2009.255 (0), 2085.338 (0),
2102.303 (0), 2177.344 (0), 2232.439 (0), 2343.487 (0), 3621.252 (0)
}
Spot: 2D-001LGC (ecoli5-6)
pI: 5.18 Mw: 32691
%vol: 0.241066 %od: 0.36004
================
*TRPA_ECOLI*
accession n°: P0A877
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 874.557 (0), 972.647 (0), 1098.73 (0), 1146.829 (0),
1349.877 (0), 1359.779 (0), 1543.024 (0), 2009.255 (0), 2085.338 (0),
2102.303 (0), 2177.344 (0), 2232.439 (0), 2343.487 (0), 3621.252 (0)
}
[Click to access protein entry]
Spot: 2D-001KZ7 (ecoli5-6)
pI: 5.71 Mw: 52000
%vol: 0.134203 %od: 0.41502
================
*DPPA_ECOLI*
accession n°: P23847
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 937.348 (0), 947.357 (0), 971.324 (0), 1050.407 (0),
1111.352 (0), 1327.528 (0), 1526.468 (0), 1890.547 (0), 2059.728 (0),
2401.735 (0), 2406.716 (0), 2642.877 (0), 3885.08 (0)
}
Spot: 2D-001KZ7 (ecoli5-6)
pI: 5.71 Mw: 52000
%vol: 0.134203 %od: 0.41502
================
*DPPA_ECOLI*
accession n°: P23847
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 937.348 (0), 947.357 (0), 971.324 (0), 1050.407 (0),
1111.352 (0), 1327.528 (0), 1526.468 (0), 1890.547 (0), 2059.728 (0),
2401.735 (0), 2406.716 (0), 2642.877 (0), 3885.08 (0)
}
[Click to access protein entry]
Spot: 2D-001KYP (ecoli5-6)
pI: 5.34 Mw: 53476
%vol: 0.102032 %od: 0.103901
================
*AROA_ECOLI*
accession n°: P0A6D3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 865.277 (0), 1067.311 (0), 1274.429 (0), 1375.483 (0),
1386.382 (0), 1460.462 (0), 1473.363 (0), 1563.427 (0), 1601.453 (0),
1635.484 (0), 1791.359 (0) }
Spot: 2D-001KYP (ecoli5-6)
pI: 5.34 Mw: 53476
%vol: 0.102032 %od: 0.103901
================
*AROA_ECOLI*
accession n°: P0A6D3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 865.277 (0), 1067.311 (0), 1274.429 (0), 1375.483 (0),
1386.382 (0), 1460.462 (0), 1473.363 (0), 1563.427 (0), 1601.453 (0),
1635.484 (0), 1791.359 (0) }
[Click to access protein entry]
Spot: 2D-001LJ2 (ecoli5-6)
pI: 5.73 Mw: 29525
%vol: 0.171032 %od: 0.250126
================
*SUCD_ECOLI*
accession n°: P0AGE9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 986.572 (0), 1068.553 (0), 1244.673 (0), 1381.792 (0),
1568.837 (0), 1866.003 (0), 1987.071 (0), 2530.08 (0), 2587.096 (0)
}
Spot: 2D-001LJ2 (ecoli5-6)
pI: 5.73 Mw: 29525
%vol: 0.171032 %od: 0.250126
================
*SUCD_ECOLI*
accession n°: P0AGE9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 986.572 (0), 1068.553 (0), 1244.673 (0), 1381.792 (0),
1568.837 (0), 1866.003 (0), 1987.071 (0), 2530.08 (0), 2587.096 (0)
}
[Click to access protein entry]
Spot: 2D-001LGL (ecoli5-6)
pI: 5.47 Mw: 32502
%vol: 0.194319 %od: 0.299908
================
*NFNB_ECOLI*
accession n°: P38489
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 923.475 (0), 939.455 (0), 1051.573 (0), 1067.54 (0),
1216.623 (0), 1227.626 (0), 1284.743 (0), 1330.629 (0), 1370.822 (0),
2566.264 (0) }
Spot: 2D-001LGL (ecoli5-6)
pI: 5.47 Mw: 32502
%vol: 0.194319 %od: 0.299908
================
*NFNB_ECOLI*
accession n°: P38489
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 923.475 (0), 939.455 (0), 1051.573 (0), 1067.54 (0),
1216.623 (0), 1227.626 (0), 1284.743 (0), 1330.629 (0), 1370.822 (0),
2566.264 (0) }
[Click to access protein entry]
Spot: 2D-001LSR (ecoli5-6)
pI: 5.44 Mw: 14684
%vol: 0.0715003 %od: 0.0687032
================
*NUSG_ECOLI*
accession n°: P0AFG0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 864.74 (0), 1105.967 (0), 1178.906 (0), 1194.9 (0),
1266.076 (0), 1463.123 (0), 1534.173 (0), 1848.43 (0), 1864.397 (0)
}
Spot: 2D-001LSR (ecoli5-6)
pI: 5.44 Mw: 14684
%vol: 0.0715003 %od: 0.0687032
================
*NUSG_ECOLI*
accession n°: P0AFG0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 864.74 (0), 1105.967 (0), 1178.906 (0), 1194.9 (0),
1266.076 (0), 1463.123 (0), 1534.173 (0), 1848.43 (0), 1864.397 (0)
}
[Click to access protein entry]
Spot: 2D-001KXJ (ecoli5-6)
pI: 5.25 Mw: 59333
%vol: 0.0605469 %od: 0.0587151
================
*LIVK_ECOLI*
accession n°: P04816
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 892.485 (0), 920.424 (0), 1021.504 (0), 1129.594 (0),
1246.708 (0), 1480.637 (0), 1802.873 (0), 2027.89 (0), 2112.009 (0),
2445.033 (0), 2461.004 (0), 2508.195 (0), 2597.261 (0), 2636.287 (0)
}
Spot: 2D-001KXJ (ecoli5-6)
pI: 5.25 Mw: 59333
%vol: 0.0605469 %od: 0.0587151
================
*LIVK_ECOLI*
accession n°: P04816
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 892.485 (0), 920.424 (0), 1021.504 (0), 1129.594 (0),
1246.708 (0), 1480.637 (0), 1802.873 (0), 2027.89 (0), 2112.009 (0),
2445.033 (0), 2461.004 (0), 2508.195 (0), 2597.261 (0), 2636.287 (0)
}
[Click to access protein entry]
Spot: 2D-001LFI (ecoli5-6)
pI: 5.19 Mw: 33559
%vol: 0.239599 %od: 0.31012
================
*ARGD_ECOLI*
accession n°: P18335
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 886.599 (0), 1219.846 (0), 1282.882 (0), 1498.955 (0),
1618.119 (0), 1634.123 (0), 1992.243 (0), 2008.229 (0)
}
Spot: 2D-001LFI (ecoli5-6)
pI: 5.19 Mw: 33559
%vol: 0.239599 %od: 0.31012
================
*ARGD_ECOLI*
accession n°: P18335
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 886.599 (0), 1219.846 (0), 1282.882 (0), 1498.955 (0),
1618.119 (0), 1634.123 (0), 1992.243 (0), 2008.229 (0)
}
[Click to access protein entry]
Spot: 2D-001LSV (ecoli5-6)
pI: 5.19 Mw: 14514
%vol: 0.0258747 %od: 0.0269314
================
*NDK_ECOLI*
accession n°: P0A763
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1003.569 (0), 1197.626 (0), 1213.635 (0), 1288.747 (0),
1616.778 (0), 1667.89 (0), 1673.741 (0), 1961.022 (0), 2352.989 (0)
}
Spot: 2D-001LSV (ecoli5-6)
pI: 5.19 Mw: 14514
%vol: 0.0258747 %od: 0.0269314
================
*NDK_ECOLI*
accession n°: P0A763
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1003.569 (0), 1197.626 (0), 1213.635 (0), 1288.747 (0),
1616.778 (0), 1667.89 (0), 1673.741 (0), 1961.022 (0), 2352.989 (0)
}
[Click to access protein entry]
Spot: 2D-001LQC (ecoli5-6)
pI: 5.78 Mw: 17897
%vol: 0.179311 %od: 0.212426
================
*FUR_ECOLI*
accession n°: P0A9A9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1380.689 (0), 1447.768 (0), 1679.881 (0), 1695.899 (0),
2349.194 (0) }
================
*DPS_ECOLI*
accession n: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 942.642 (0), 951.751 (0), 1518.247 (0), 1677.213 (0),
1679.206 (0), 1693.222 (0) }
Spot: 2D-001LQC (ecoli5-6)
pI: 5.78 Mw: 17897
%vol: 0.179311 %od: 0.212426
================
*FUR_ECOLI*
accession n°: P0A9A9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1380.689 (0), 1447.768 (0), 1679.881 (0), 1695.899 (0),
2349.194 (0) }
================
*DPS_ECOLI*
accession n: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 942.642 (0), 951.751 (0), 1518.247 (0), 1677.213 (0),
1679.206 (0), 1693.222 (0) }
[Click to access protein entry]
Spot: 2D-001L7G (ecoli5-6)
pI: 5.31 Mw: 48144
%vol: 0.167927 %od: 0.217149
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.405 (0), 1489.751 (0), 1803.839 (0), 1962.019 (0),
1964.948 (0), 2117.178 (0), 2729.45 (0), 2745.395 (0)
}
Spot: 2D-001L7G (ecoli5-6)
pI: 5.31 Mw: 48144
%vol: 0.167927 %od: 0.217149
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.405 (0), 1489.751 (0), 1803.839 (0), 1962.019 (0),
1964.948 (0), 2117.178 (0), 2729.45 (0), 2745.395 (0)
}
[Click to access protein entry]
Spot: 2D-001L6H (ecoli5-6)
pI: 5.34 Mw: 50034
%vol: 0.0898714 %od: 0.102348
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.694 (0), 1171.76 (0), 1203.627 (0), 1214.737 (0),
1218.674 (0), 1376.782 (0), 1489.988 (0), 1575.116 (0), 1768.959 (0),
1796.115 (0), 1804.059 (0), 1962.214 (0), 1965.152 (0), 2117.375 (0)
}
Spot: 2D-001L6H (ecoli5-6)
pI: 5.34 Mw: 50034
%vol: 0.0898714 %od: 0.102348
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.694 (0), 1171.76 (0), 1203.627 (0), 1214.737 (0),
1218.674 (0), 1376.782 (0), 1489.988 (0), 1575.116 (0), 1768.959 (0),
1796.115 (0), 1804.059 (0), 1962.214 (0), 1965.152 (0), 2117.375 (0)
}
[Click to access protein entry]
Spot: 2D-001LS6 (ecoli5-6)
pI: 5.63 Mw: 15384
%vol: 0.104447 %od: 0.0945373
================
*IBPA_ECOLI*
accession n°: P0C054
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1124.574 (0), 1140.567 (0), 1213.636 (0), 1226.676 (0),
1354.781 (0), 1659.936 (0) }
Spot: 2D-001LS6 (ecoli5-6)
pI: 5.63 Mw: 15384
%vol: 0.104447 %od: 0.0945373
================
*IBPA_ECOLI*
accession n°: P0C054
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1124.574 (0), 1140.567 (0), 1213.636 (0), 1226.676 (0),
1354.781 (0), 1659.936 (0) }
[Click to access protein entry]
Spot: 2D-001L5A (ecoli5-6)
pI: 5.20 Mw: 50339
%vol: 0.218468 %od: 0.447979
================
*DHAS_ECOLI*
accession n°: P0A9Q9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 948.522 (0), 1077.591 (0), 1093.556 (0), 1683.976 (0),
1742.981 (0), 1825.885 (0), 2090.113 (0), 2270.114 (0), 2852.461 (0),
4118.99 (0) }
Spot: 2D-001L5A (ecoli5-6)
pI: 5.20 Mw: 50339
%vol: 0.218468 %od: 0.447979
================
*DHAS_ECOLI*
accession n°: P0A9Q9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 948.522 (0), 1077.591 (0), 1093.556 (0), 1683.976 (0),
1742.981 (0), 1825.885 (0), 2090.113 (0), 2270.114 (0), 2852.461 (0),
4118.99 (0) }
[Click to access protein entry]
Spot: 2D-001LMN (ecoli5-6)
pI: 5.60 Mw: 24224
%vol: 0.0972025 %od: 0.0760007
================
*CLPP_ECOLI*
accession n°: P0A6G7
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 887.576 (0), 893.578 (0), 1049.696 (0), 2100.357 (0),
2217.505 (0), 2434.612 (0), 2450.563 (0) }
Spot: 2D-001LMN (ecoli5-6)
pI: 5.60 Mw: 24224
%vol: 0.0972025 %od: 0.0760007
================
*CLPP_ECOLI*
accession n°: P0A6G7
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 887.576 (0), 893.578 (0), 1049.696 (0), 2100.357 (0),
2217.505 (0), 2434.612 (0), 2450.563 (0) }
[Click to access protein entry]
Spot: 2D-001L7Q (ecoli5-6)
pI: 5.33 Mw: 47448
%vol: 0.178708 %od: 0.160437
================
*YAET_ECOLI*
accession n°: P0A940
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 889.52 (0), 973.606 (0), 987.656 (0), 1114.684 (0),
1220.726 (0), 1277.743 (0), 1283.905 (0), 1339.814 (0), 1471.874 (0),
1520.978 (0), 1649.946 (0), 1693.053 (0), 1700.042 (0), 2239.361 (0)
}
================
*EFTU2_ECOLI*
accession n: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.705 (0), 1171.768 (0), 1203.657 (0), 1214.745 (0),
1768.965 (0), 1796.141 (0), 1804.088 (0), 1962.244 (0), 1965.174 (0),
2117.39 (0) }
Spot: 2D-001L7Q (ecoli5-6)
pI: 5.33 Mw: 47448
%vol: 0.178708 %od: 0.160437
================
*YAET_ECOLI*
accession n°: P0A940
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 889.52 (0), 973.606 (0), 987.656 (0), 1114.684 (0),
1220.726 (0), 1277.743 (0), 1283.905 (0), 1339.814 (0), 1471.874 (0),
1520.978 (0), 1649.946 (0), 1693.053 (0), 1700.042 (0), 2239.361 (0)
}
================
*EFTU2_ECOLI*
accession n: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.705 (0), 1171.768 (0), 1203.657 (0), 1214.745 (0),
1768.965 (0), 1796.141 (0), 1804.088 (0), 1962.244 (0), 1965.174 (0),
2117.39 (0) }
[Click to access protein entry]
Spot: 2D-001LHF (ecoli5-6)
pI: 5.07 Mw: 31478
%vol: 0.197769 %od: 0.249651
================
*ARTI_ECOLI*
accession n°: P30859
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1045.656 (0), 1203.684 (0), 1268.805 (0), 1675.07 (0),
1712.13 (0), 1863.1 (0), 2300.463 (0) }
Spot: 2D-001LHF (ecoli5-6)
pI: 5.07 Mw: 31478
%vol: 0.197769 %od: 0.249651
================
*ARTI_ECOLI*
accession n°: P30859
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1045.656 (0), 1203.684 (0), 1268.805 (0), 1675.07 (0),
1712.13 (0), 1863.1 (0), 2300.463 (0) }
[Click to access protein entry]
Spot: 2D-001LPG (ecoli5-6)
pI: 5.46 Mw: 19137
%vol: 0.0373459 %od: 0.0398462
================
*SSB_ECOLI*
accession n°: P0AGE0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 985.62 (0), 1092.676 (0), 1150.822 (0), 1220.804 (0),
1249.884 (0), 1378.002 (0), 1509.116 (0), 2044.332 (0)
}
Spot: 2D-001LPG (ecoli5-6)
pI: 5.46 Mw: 19137
%vol: 0.0373459 %od: 0.0398462
================
*SSB_ECOLI*
accession n°: P0AGE0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 985.62 (0), 1092.676 (0), 1150.822 (0), 1220.804 (0),
1249.884 (0), 1378.002 (0), 1509.116 (0), 2044.332 (0)
}
[Click to access protein entry]
Spot: 2D-001LD0 (ecoli5-6)
pI: 5.80 Mw: 37049
%vol: 0.0954775 %od: 0.105582
================
*EX3_ECOLI*
accession n°: P09030
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1009.473 (0), 1020.454 (0), 1037.476 (0), 1058.547 (0),
1166.605 (0), 1220.524 (0), 1324.66 (0), 1325.67 (0), 1364.714 (0),
1376.649 (0), 1390.781 (0), 1858.886 (0), 2158.093 (0)
}
Spot: 2D-001LD0 (ecoli5-6)
pI: 5.80 Mw: 37049
%vol: 0.0954775 %od: 0.105582
================
*EX3_ECOLI*
accession n°: P09030
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1009.473 (0), 1020.454 (0), 1037.476 (0), 1058.547 (0),
1166.605 (0), 1220.524 (0), 1324.66 (0), 1325.67 (0), 1364.714 (0),
1376.649 (0), 1390.781 (0), 1858.886 (0), 2158.093 (0)
}
[Click to access protein entry]
Spot: 2D-001LEJ (ecoli5-6)
pI: 5.44 Mw: 34853
%vol: 0.175344 %od: 0.189204
================
*CLPB_ECOLI*
accession n°: P63284
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 915.399 (0), 955.354 (0), 971.372 (0), 1079.384 (0),
1095.377 (0), 1145.348 (0), 1187.473 (0), 1210.441 (0), 1226.429 (0),
1286.568 (0), 1560.698 (0), 1655.694 (0), 1732.586 (0), 1748.565 (0),
1921.823 (0), 1989.81 (0), 2005.79 (0), 2125.913 (0), 2283.991 (0),
2296.898 (0), 2684.143 (0) }
Spot: 2D-001LEJ (ecoli5-6)
pI: 5.44 Mw: 34853
%vol: 0.175344 %od: 0.189204
================
*CLPB_ECOLI*
accession n°: P63284
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 915.399 (0), 955.354 (0), 971.372 (0), 1079.384 (0),
1095.377 (0), 1145.348 (0), 1187.473 (0), 1210.441 (0), 1226.429 (0),
1286.568 (0), 1560.698 (0), 1655.694 (0), 1732.586 (0), 1748.565 (0),
1921.823 (0), 1989.81 (0), 2005.79 (0), 2125.913 (0), 2283.991 (0),
2296.898 (0), 2684.143 (0) }
[Click to access protein entry]
Spot: 2D-001L39 (ecoli5-6)
pI: 5.00 Mw: 50698
%vol: 0.198976 %od: 0.241261
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 928.562 (0), 950.51 (0), 1189.619 (0), 1248.607 (0),
1255.651 (0), 1271.616 (0), 1459.83 (0), 1492.886 (0), 1546.832 (0),
1562.848 (0), 1605.893 (0), 1826.912 (0), 1842.875 (0), 1928.93 (0),
2075.116 (0), 2118.046 (0), 2776.22 (0) }
Spot: 2D-001L39 (ecoli5-6)
pI: 5.00 Mw: 50698
%vol: 0.198976 %od: 0.241261
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 928.562 (0), 950.51 (0), 1189.619 (0), 1248.607 (0),
1255.651 (0), 1271.616 (0), 1459.83 (0), 1492.886 (0), 1546.832 (0),
1562.848 (0), 1605.893 (0), 1826.912 (0), 1842.875 (0), 1928.93 (0),
2075.116 (0), 2118.046 (0), 2776.22 (0) }
[Click to access protein entry]
Spot: 2D-001L3N (ecoli5-6)
pI: 5.16 Mw: 50629
%vol: 0.197855 %od: 0.347725
================
*MANA_ECOLI*
accession n°: P00946
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1097.537 (0), 1242.662 (0), 1253.676 (0), 1344.606 (0),
1373.725 (0), 1715.85 (0), 2599.425 (0), 2989.391 (0), 3270.714 (0)
}
Spot: 2D-001L3N (ecoli5-6)
pI: 5.16 Mw: 50629
%vol: 0.197855 %od: 0.347725
================
*MANA_ECOLI*
accession n°: P00946
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1097.537 (0), 1242.662 (0), 1253.676 (0), 1344.606 (0),
1373.725 (0), 1715.85 (0), 2599.425 (0), 2989.391 (0), 3270.714 (0)
}
[Click to access protein entry]
Spot: 2D-001L3T (ecoli5-6)
pI: 5.02 Mw: 50647
%vol: 0.110312 %od: 0.112604
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1255.753 (0), 1605.882 (0), 1826.915 (0), 1928.902 (0),
2775.948 (0), 2856.077 (0) }
================
*SERC_ECOLI*
accession n: P23721
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 968.453 (0), 1002.55 (0), 1322.923 (0), 1357.749 (0),
1442.762 (0), 1645.884 (0), 1710.873 (0), 1774.873 (0), 2019.932 (0)
}
Spot: 2D-001L3T (ecoli5-6)
pI: 5.02 Mw: 50647
%vol: 0.110312 %od: 0.112604
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1255.753 (0), 1605.882 (0), 1826.915 (0), 1928.902 (0),
2775.948 (0), 2856.077 (0) }
================
*SERC_ECOLI*
accession n: P23721
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 968.453 (0), 1002.55 (0), 1322.923 (0), 1357.749 (0),
1442.762 (0), 1645.884 (0), 1710.873 (0), 1774.873 (0), 2019.932 (0)
}
[Click to access protein entry]
Spot: 2D-001L9C (ecoli5-6)
pI: 5.78 Mw: 42978
%vol: 0.173015 %od: 0.381707
================
*CYSK_ECOLI*
accession n°: P0ABK5
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1210.565 (0), 1256.722 (0), 1283.75 (0), 1473.811 (0),
1521.782 (0), 1537.771 (0), 1553.783 (0), 1618.861 (0), 1626.926 (0),
1811.995 (0), 2141.145 (0), 2273.216 (0), 2753.494 (0)
}
Spot: 2D-001L9C (ecoli5-6)
pI: 5.78 Mw: 42978
%vol: 0.173015 %od: 0.381707
================
*CYSK_ECOLI*
accession n°: P0ABK5
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1210.565 (0), 1256.722 (0), 1283.75 (0), 1473.811 (0),
1521.782 (0), 1537.771 (0), 1553.783 (0), 1618.861 (0), 1626.926 (0),
1811.995 (0), 2141.145 (0), 2273.216 (0), 2753.494 (0)
}
[Click to access protein entry]
Spot: 2D-001KYZ (ecoli5-6)
pI: 5.48 Mw: 53050
%vol: 0.116695 %od: 0.170337
================
*GLTD_ECOLI*
accession n°: P09832
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 906.441 (0), 947.543 (0), 977.467 (0), 1038.516 (0),
1054.509 (0), 1127.56 (0), 1206.539 (0), 1271.66 (0), 1287.698 (0),
1510.78 (0), 1564.75 (0), 1610.753 (0), 1626.757 (0), 1642.754 (0),
1767.996 (0), 1788.879 (0), 1825.007 (0), 1953.141 (0), 2014.014 (0),
2272.117 (0), 2455.135 (0) }
Spot: 2D-001KYZ (ecoli5-6)
pI: 5.48 Mw: 53050
%vol: 0.116695 %od: 0.170337
================
*GLTD_ECOLI*
accession n°: P09832
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 906.441 (0), 947.543 (0), 977.467 (0), 1038.516 (0),
1054.509 (0), 1127.56 (0), 1206.539 (0), 1271.66 (0), 1287.698 (0),
1510.78 (0), 1564.75 (0), 1610.753 (0), 1626.757 (0), 1642.754 (0),
1767.996 (0), 1788.879 (0), 1825.007 (0), 1953.141 (0), 2014.014 (0),
2272.117 (0), 2455.135 (0) }
[Click to access protein entry]
Spot: 2D-001LMY (ecoli5-6)
pI: 5.41 Mw: 23874
%vol: 0.0815051 %od: 0.0890387
================
*SSB_ECOLI*
accession n°: P0AGE0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 985.643 (0), 1092.685 (0), 1150.844 (0), 1220.828 (0),
1249.896 (0), 1378.028 (0), 1509.134 (0) }
Spot: 2D-001LMY (ecoli5-6)
pI: 5.41 Mw: 23874
%vol: 0.0815051 %od: 0.0890387
================
*SSB_ECOLI*
accession n°: P0AGE0
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 985.643 (0), 1092.685 (0), 1150.844 (0), 1220.828 (0),
1249.896 (0), 1378.028 (0), 1509.134 (0) }
[Click to access protein entry]
Spot: 2D-001LHX (ecoli5-6)
pI: 5.67 Mw: 30753
%vol: 0.175172 %od: 0.346788
================
*LIVJ_ECOLI*
accession n°: P0AD96
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 862.423 (0), 908.378 (0), 1021.391 (0), 1361.494 (0),
1638.597 (0), 1766.684 (0), 2400.761 (0), 2416.705 (0)
}
Spot: 2D-001LHX (ecoli5-6)
pI: 5.67 Mw: 30753
%vol: 0.175172 %od: 0.346788
================
*LIVJ_ECOLI*
accession n°: P0AD96
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 862.423 (0), 908.378 (0), 1021.391 (0), 1361.494 (0),
1638.597 (0), 1766.684 (0), 2400.761 (0), 2416.705 (0)
}
[Click to access protein entry]
Spot: 2D-001L6U (ecoli5-6)
pI: 5.56 Mw: 49421
%vol: 0.193629 %od: 0.412127
================
*AAT_ECOLI*
accession n°: P00509
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1012.489 (0), 1088.59 (0), 1129.62 (0), 1178.622 (0),
1450.748 (0), 2438.173 (0) }
================
*ALF_ECOLI*
accession n: P0AB71
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 934.591 (0), 950.583 (0), 953.573 (0), 1320.797 (0),
1502.947 (0), 1878.221 (0), 2591.534 (0), 2719.66 (0)
}
Spot: 2D-001L6U (ecoli5-6)
pI: 5.56 Mw: 49421
%vol: 0.193629 %od: 0.412127
================
*AAT_ECOLI*
accession n°: P00509
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1012.489 (0), 1088.59 (0), 1129.62 (0), 1178.622 (0),
1450.748 (0), 2438.173 (0) }
================
*ALF_ECOLI*
accession n: P0AB71
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 934.591 (0), 950.583 (0), 953.573 (0), 1320.797 (0),
1502.947 (0), 1878.221 (0), 2591.534 (0), 2719.66 (0)
}
[Click to access protein entry]
Spot: 2D-001LKA (ecoli5-6)
pI: 5.51 Mw: 28018
%vol: 0.140068 %od: 0.178026
================
*WRBA_ECOLI*
accession n°: P0A8G6
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.651 (0), 1059.693 (0), 1257.934 (0), 1297.984 (0),
1538.085 (0), 1588.182 (0), 2676.94 (0) }
Spot: 2D-001LKA (ecoli5-6)
pI: 5.51 Mw: 28018
%vol: 0.140068 %od: 0.178026
================
*WRBA_ECOLI*
accession n°: P0A8G6
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.651 (0), 1059.693 (0), 1257.934 (0), 1297.984 (0),
1538.085 (0), 1588.182 (0), 2676.94 (0) }
[Click to access protein entry]
Spot: 2D-001LPP (ecoli5-6)
pI: 5.74 Mw: 18915
%vol: 0.115315 %od: 0.127603
================
*MOAB_ECOLI*
accession n°: P0AEZ9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 986.695 (0), 1168.683 (0), 1264.781 (0), 1534.887 (0),
1698.932 (0) }
Spot: 2D-001LPP (ecoli5-6)
pI: 5.74 Mw: 18915
%vol: 0.115315 %od: 0.127603
================
*MOAB_ECOLI*
accession n°: P0AEZ9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 986.695 (0), 1168.683 (0), 1264.781 (0), 1534.887 (0),
1698.932 (0) }
[Click to access protein entry]
Spot: 2D-001LTD (ecoli5-6)
pI: 5.45 Mw: 13773
%vol: 0.184573 %od: 0.380574
================
*ATPE_ECOLI*
accession n°: P0A6E6
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 903.636 (0), 2415.594 (0), 2543.732 (0), 3019.069 (0),
3034.957 (0) }
Spot: 2D-001LTD (ecoli5-6)
pI: 5.45 Mw: 13773
%vol: 0.184573 %od: 0.380574
================
*ATPE_ECOLI*
accession n°: P0A6E6
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 903.636 (0), 2415.594 (0), 2543.732 (0), 3019.069 (0),
3034.957 (0) }
[Click to access protein entry]
Spot: 2D-001LSW (ecoli5-6)
pI: 5.41 Mw: 14304
%vol: 0.17198 %od: 0.439112
================
*IVY_ECOLI*
accession n°: P0AD59
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.648 (0), 979.646 (0), 1230.829 (0), 1246.818 (0),
1571.036 (0), 1587.033 (0), 1659.198 (0), 2278.558 (0)
}
Spot: 2D-001LSW (ecoli5-6)
pI: 5.41 Mw: 14304
%vol: 0.17198 %od: 0.439112
================
*IVY_ECOLI*
accession n°: P0AD59
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.648 (0), 979.646 (0), 1230.829 (0), 1246.818 (0),
1571.036 (0), 1587.033 (0), 1659.198 (0), 2278.558 (0)
}
[Click to access protein entry]
Spot: 2D-001L6J (ecoli5-6)
pI: 5.43 Mw: 50034
%vol: 0.119541 %od: 0.110291
================
*ALDA_ECOLI*
accession n°: P25553
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 974.59 (0), 999.502 (0), 1011.554 (0), 1111.541 (0),
1569.825 (0), 2233.091 (0) }
Spot: 2D-001L6J (ecoli5-6)
pI: 5.43 Mw: 50034
%vol: 0.119541 %od: 0.110291
================
*ALDA_ECOLI*
accession n°: P25553
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 974.59 (0), 999.502 (0), 1011.554 (0), 1111.541 (0),
1569.825 (0), 2233.091 (0) }
[Click to access protein entry]
Spot: 2D-001LL3 (ecoli5-6)
pI: 5.24 Mw: 27057
%vol: 0.049852 %od: 0.0298487
================
*HIS5_ECOLI*
accession n°: P60595
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 877.627 (0), 904.613 (0), 1162.833 (0), 1427.015 (0),
1508.932 (0), 1774.206 (0), 1864.215 (0), 2185.464 (0)
}
Spot: 2D-001LL3 (ecoli5-6)
pI: 5.24 Mw: 27057
%vol: 0.049852 %od: 0.0298487
================
*HIS5_ECOLI*
accession n°: P60595
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 877.627 (0), 904.613 (0), 1162.833 (0), 1427.015 (0),
1508.932 (0), 1774.206 (0), 1864.215 (0), 2185.464 (0)
}
[Click to access protein entry]
Spot: 2D-001L85 (ecoli5-6)
pI: 5.43 Mw: 45820
%vol: 0.155938 %od: 0.330233
================
*K6PF1_ECOLI*
accession n°: P0A796
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 865.593 (0), 947.599 (0), 994.732 (0), 1084.684 (0),
1100.665 (0), 1641.08 (0), 1657.082 (0), 1700.126 (0), 1716.091 (0)
}
================
*NADE_ECOLI*
accession n: P18843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 911.537 (0), 1114.627 (0), 1235.651 (0), 1440.689 (0),
1576.835 (0), 1638.887 (0) }
Spot: 2D-001L85 (ecoli5-6)
pI: 5.43 Mw: 45820
%vol: 0.155938 %od: 0.330233
================
*K6PF1_ECOLI*
accession n°: P0A796
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 865.593 (0), 947.599 (0), 994.732 (0), 1084.684 (0),
1100.665 (0), 1641.08 (0), 1657.082 (0), 1700.126 (0), 1716.091 (0)
}
================
*NADE_ECOLI*
accession n: P18843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 911.537 (0), 1114.627 (0), 1235.651 (0), 1440.689 (0),
1576.835 (0), 1638.887 (0) }
[Click to access protein entry]
Spot: 2D-001LUG (ecoli5-6)
pI: 5.78 Mw: 12732
%vol: 0.156973 %od: 0.211198
================
*GLNK_ECOLI*
accession n°: P0AC55
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 845.49 (0), 975.588 (0), 1101.568 (0), 1224.583 (0),
1744.755 (0), 1769.868 (0) }
Spot: 2D-001LUG (ecoli5-6)
pI: 5.78 Mw: 12732
%vol: 0.156973 %od: 0.211198
================
*GLNK_ECOLI*
accession n°: P0AC55
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 845.49 (0), 975.588 (0), 1101.568 (0), 1224.583 (0),
1744.755 (0), 1769.868 (0) }
[Click to access protein entry]
Spot: 2D-001L0Q (ecoli5-6)
pI: 5.27 Mw: 51301
%vol: 0.302216 %od: 1.18916
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 888.415 (0), 928.503 (0), 950.43 (0), 1145.492 (0),
1189.527 (0), 1248.519 (0), 1255.537 (0), 1264.527 (0), 1271.535 (0),
1459.725 (0), 1492.729 (0), 1562.731 (0), 1605.769 (0), 1674.784 (0),
1750.681 (0), 1928.782 (0), 2117.906 (0), 2856.118 (0)
}
Spot: 2D-001L0Q (ecoli5-6)
pI: 5.27 Mw: 51301
%vol: 0.302216 %od: 1.18916
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 888.415 (0), 928.503 (0), 950.43 (0), 1145.492 (0),
1189.527 (0), 1248.519 (0), 1255.537 (0), 1264.527 (0), 1271.535 (0),
1459.725 (0), 1492.729 (0), 1562.731 (0), 1605.769 (0), 1674.784 (0),
1750.681 (0), 1928.782 (0), 2117.906 (0), 2856.118 (0)
}
[Click to access protein entry]
Spot: 2D-001LGQ (ecoli5-6)
pI: 5.82 Mw: 32032
%vol: 0.206825 %od: 0.620496
================
*GPMA_ECOLI*
accession n°: P62707
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 985.673 (0), 1100.853 (0), 1150.945 (0), 1159.912 (0),
1316.052 (0), 1328.016 (0), 1356.934 (0), 1501.194 (0), 1571.25 (0),
1756.243 (0), 2110.557 (0) }
Spot: 2D-001LGQ (ecoli5-6)
pI: 5.82 Mw: 32032
%vol: 0.206825 %od: 0.620496
================
*GPMA_ECOLI*
accession n°: P62707
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 985.673 (0), 1100.853 (0), 1150.945 (0), 1159.912 (0),
1316.052 (0), 1328.016 (0), 1356.934 (0), 1501.194 (0), 1571.25 (0),
1756.243 (0), 2110.557 (0) }
[Click to access protein entry]
Spot: 2D-001L2D (ecoli5-6)
pI: 5.32 Mw: 50990
%vol: 0.17612 %od: 0.241993
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.699 (0), 1171.772 (0), 1203.675 (0), 1218.707 (0),
1233.768 (0), 1689.98 (0), 1768.981 (0), 1796.163 (0), 1804.075 (0),
1962.231 (0), 1965.178 (0), 2117.363 (0) }
Spot: 2D-001L2D (ecoli5-6)
pI: 5.32 Mw: 50990
%vol: 0.17612 %od: 0.241993
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.699 (0), 1171.772 (0), 1203.675 (0), 1218.707 (0),
1233.768 (0), 1689.98 (0), 1768.981 (0), 1796.163 (0), 1804.075 (0),
1962.231 (0), 1965.178 (0), 2117.363 (0) }
[Click to access protein entry]
Spot: 2D-001L27 (ecoli5-6)
pI: 5.23 Mw: 50990
%vol: 0.218986 %od: 0.266145
================
*AROA_ECOLI*
accession n°: P0A6D3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1067.376 (0), 1092.374 (0), 1179.437 (0), 1274.475 (0),
1375.566 (0), 1386.476 (0), 1460.544 (0), 1473.488 (0), 1552.547 (0),
1563.521 (0), 1601.556 (0), 1635.604 (0), 1791.459 (0)
}
Spot: 2D-001L27 (ecoli5-6)
pI: 5.23 Mw: 50990
%vol: 0.218986 %od: 0.266145
================
*AROA_ECOLI*
accession n°: P0A6D3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1067.376 (0), 1092.374 (0), 1179.437 (0), 1274.475 (0),
1375.566 (0), 1386.476 (0), 1460.544 (0), 1473.488 (0), 1552.547 (0),
1563.521 (0), 1601.556 (0), 1635.604 (0), 1791.459 (0)
}
[Click to access protein entry]
Spot: 2D-001L0V (ecoli5-6)
pI: 5.16 Mw: 51319
%vol: 0.301009 %od: 0.949717
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 950.475 (0), 1189.571 (0), 1255.612 (0), 1271.597 (0),
1459.808 (0), 1492.843 (0), 1562.837 (0), 1605.869 (0), 1750.766 (0),
1826.889 (0), 1842.893 (0), 1928.926 (0), 2118.06 (0), 2776.217 (0),
2856.331 (0), 2872.293 (0) }
Spot: 2D-001L0V (ecoli5-6)
pI: 5.16 Mw: 51319
%vol: 0.301009 %od: 0.949717
================
*ENO_ECOLI*
accession n°: P0A6P9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 950.475 (0), 1189.571 (0), 1255.612 (0), 1271.597 (0),
1459.808 (0), 1492.843 (0), 1562.837 (0), 1605.869 (0), 1750.766 (0),
1826.889 (0), 1842.893 (0), 1928.926 (0), 2118.06 (0), 2776.217 (0),
2856.331 (0), 2872.293 (0) }
[Click to access protein entry]
Spot: 2D-001L13 (ecoli5-6)
pI: 5.39 Mw: 51249
%vol: 0.149642 %od: 0.335583
================
*PROA_ECOLI*
accession n°: P07004
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 918.618 (0), 1020.656 (0), 1036.669 (0), 1107.692 (0),
1193.781 (0), 1275.825 (0), 1437.96 (0), 1464.904 (0), 1683.982 (0),
1741.019 (0), 1858.13 (0), 1961.219 (0), 2569.527 (0)
}
================
*EFTU2_ECOLI*
accession n: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.686 (0), 1233.741 (0), 1768.93 (0), 1781.083 (0),
1796.085 (0), 1804.033 (0), 1962.177 (0), 1965.13 (0), 2117.35 (0)
}
Spot: 2D-001L13 (ecoli5-6)
pI: 5.39 Mw: 51249
%vol: 0.149642 %od: 0.335583
================
*PROA_ECOLI*
accession n°: P07004
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 918.618 (0), 1020.656 (0), 1036.669 (0), 1107.692 (0),
1193.781 (0), 1275.825 (0), 1437.96 (0), 1464.904 (0), 1683.982 (0),
1741.019 (0), 1858.13 (0), 1961.219 (0), 2569.527 (0)
}
================
*EFTU2_ECOLI*
accession n: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.686 (0), 1233.741 (0), 1768.93 (0), 1781.083 (0),
1796.085 (0), 1804.033 (0), 1962.177 (0), 1965.13 (0), 2117.35 (0)
}
[Click to access protein entry]
Spot: 2D-001LW8 (ecoli5-6)
pI: 5.77 Mw: 11202
%vol: 0.177845 %od: 0.293456
================
*YGIN_ECOLI*
accession n°: P0ADU2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 988.48 (0), 1019.535 (0), 1045.564 (0), 1061.56 (0),
1810.932 (0) }
Spot: 2D-001LW8 (ecoli5-6)
pI: 5.77 Mw: 11202
%vol: 0.177845 %od: 0.293456
================
*YGIN_ECOLI*
accession n°: P0ADU2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 988.48 (0), 1019.535 (0), 1045.564 (0), 1061.56 (0),
1810.932 (0) }
[Click to access protein entry]
Spot: 2D-001LT4 (ecoli5-6)
pI: 5.23 Mw: 14346
%vol: 0.18785 %od: 0.401586
================
*OMPA_ECOLI*
accession n°: P0A910
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 872.616 (0), 1083.635 (0), 1157.717 (0), 1214.735 (0),
1222.769 (0), 1280.769 (0), 1370.838 (0), 1378.901 (0), 1409.804 (0),
2600.508 (0) }
Spot: 2D-001LT4 (ecoli5-6)
pI: 5.23 Mw: 14346
%vol: 0.18785 %od: 0.401586
================
*OMPA_ECOLI*
accession n°: P0A910
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 872.616 (0), 1083.635 (0), 1157.717 (0), 1214.735 (0),
1222.769 (0), 1280.769 (0), 1370.838 (0), 1378.901 (0), 1409.804 (0),
2600.508 (0) }
[Click to access protein entry]
Spot: 2D-001LI0 (ecoli5-6)
pI: 5.42 Mw: 30843
%vol: 0.152488 %od: 0.132151
================
*NUOB_ECOLI*
accession n°: P0AFC7
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 899.502 (0), 915.522 (0), 1104.639 (0), 1120.642 (0),
1136.666 (0), 1152.672 (0), 1632.019 (0), 1641.979 (0)
}
Spot: 2D-001LI0 (ecoli5-6)
pI: 5.42 Mw: 30843
%vol: 0.152488 %od: 0.132151
================
*NUOB_ECOLI*
accession n°: P0AFC7
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 899.502 (0), 915.522 (0), 1104.639 (0), 1120.642 (0),
1136.666 (0), 1152.672 (0), 1632.019 (0), 1641.979 (0)
}
[Click to access protein entry]
Spot: 2D-001LQT (ecoli5-6)
pI: 5.78 Mw: 17233
%vol: 0.0743467 %od: 0.084514
================
*FUR_ECOLI*
accession n°: P0A9A9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1380.684 (0), 1447.778 (0), 1679.899 (0), 1695.887 (0),
2165.99 (0), 2349.217 (0) }
Spot: 2D-001LQT (ecoli5-6)
pI: 5.78 Mw: 17233
%vol: 0.0743467 %od: 0.084514
================
*FUR_ECOLI*
accession n°: P0A9A9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1380.684 (0), 1447.778 (0), 1679.899 (0), 1695.887 (0),
2165.99 (0), 2349.217 (0) }
[Click to access protein entry]
Spot: 2D-001L3W (ecoli5-6)
pI: 5.09 Mw: 50544
%vol: 0.288589 %od: 1.16876
================
*LIVJ_ECOLI*
accession n°: P0AD96
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 862.449 (0), 908.41 (0), 1021.427 (0), 1361.551 (0),
1638.668 (0), 1707.623 (0), 1762.67 (0), 1766.758 (0), 1784.785 (0),
2069.772 (0), 2400.94 (0), 2416.912 (0), 2482.919 (0), 2528.083 (0)
}
================
*MALE_ECOLI*
accession n: P0AEX9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 959.53 (0), 1267.698 (0), 1766.898 (0), 1891.007 (0),
2139.147 (0) }
Spot: 2D-001L3W (ecoli5-6)
pI: 5.09 Mw: 50544
%vol: 0.288589 %od: 1.16876
================
*LIVJ_ECOLI*
accession n°: P0AD96
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 862.449 (0), 908.41 (0), 1021.427 (0), 1361.551 (0),
1638.668 (0), 1707.623 (0), 1762.67 (0), 1766.758 (0), 1784.785 (0),
2069.772 (0), 2400.94 (0), 2416.912 (0), 2482.919 (0), 2528.083 (0)
}
================
*MALE_ECOLI*
accession n: P0AEX9
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 959.53 (0), 1267.698 (0), 1766.898 (0), 1891.007 (0),
2139.147 (0) }
[Click to access protein entry]
Spot: 2D-001LUK (ecoli5-6)
pI: 5.33 Mw: 12548
%vol: 0.0591666 %od: 0.0767456
================
*IVY_ECOLI*
accession n°: P0AD59
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.635 (0), 979.645 (0), 1230.825 (0), 1246.814 (0),
1587.016 (0), 2278.55 (0) }
Spot: 2D-001LUK (ecoli5-6)
pI: 5.33 Mw: 12548
%vol: 0.0591666 %od: 0.0767456
================
*IVY_ECOLI*
accession n°: P0AD59
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.635 (0), 979.645 (0), 1230.825 (0), 1246.814 (0),
1587.016 (0), 2278.55 (0) }
[Click to access protein entry]
Spot: 2D-001LM6 (ecoli5-6)
pI: 5.49 Mw: 25158
%vol: 0.185521 %od: 0.474647
================
*SODF_ECOLI*
accession n°: P0AGD3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.696 (0), 1235.893 (0), 1294.912 (0), 1631.177 (0),
1695.236 (0), 2015.391 (0) }
Spot: 2D-001LM6 (ecoli5-6)
pI: 5.49 Mw: 25158
%vol: 0.185521 %od: 0.474647
================
*SODF_ECOLI*
accession n°: P0AGD3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.696 (0), 1235.893 (0), 1294.912 (0), 1631.177 (0),
1695.236 (0), 2015.391 (0) }
[Click to access protein entry]
Spot: 2D-001LA1 (ecoli5-6)
pI: 5.20 Mw: 41503
%vol: 0.0443319 %od: 0.0388624
================
*AHPF_ECOLI*
accession n°: P35340
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 929.545 (0), 930.566 (0), 1186.742 (0), 1330.817 (0),
1404.86 (0), 1713.053 (0), 1774.975 (0), 2006.097 (0), 2016.168 (0),
2174.378 (0), 2208.271 (0) }
Spot: 2D-001LA1 (ecoli5-6)
pI: 5.20 Mw: 41503
%vol: 0.0443319 %od: 0.0388624
================
*AHPF_ECOLI*
accession n°: P35340
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 929.545 (0), 930.566 (0), 1186.742 (0), 1330.817 (0),
1404.86 (0), 1713.053 (0), 1774.975 (0), 2006.097 (0), 2016.168 (0),
2174.378 (0), 2208.271 (0) }
[Click to access protein entry]
Spot: 2D-001LA5 (ecoli5-6)
pI: 5.57 Mw: 40903
%vol: 0.230975 %od: 0.887471
================
*MDH_ECOLI*
accession n°: P61889
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 899.631 (0), 1008.641 (0), 1149.793 (0), 1193.776 (0),
1234.771 (0), 1247.881 (0), 1276.824 (0), 1405.839 (0), 1414.923 (0),
1421.853 (0), 1735.262 (0), 2366.454 (0), 2399.35 (0), 2402.428 (0),
2643.668 (0), 3376.123 (0), 4312.703 (0) }
Spot: 2D-001LA5 (ecoli5-6)
pI: 5.57 Mw: 40903
%vol: 0.230975 %od: 0.887471
================
*MDH_ECOLI*
accession n°: P61889
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 899.631 (0), 1008.641 (0), 1149.793 (0), 1193.776 (0),
1234.771 (0), 1247.881 (0), 1276.824 (0), 1405.839 (0), 1414.923 (0),
1421.853 (0), 1735.262 (0), 2366.454 (0), 2399.35 (0), 2402.428 (0),
2643.668 (0), 3376.123 (0), 4312.703 (0) }
[Click to access protein entry]
Spot: 2D-001LCK (ecoli5-6)
pI: 5.33 Mw: 37266
%vol: 0.194923 %od: 0.386849
================
*KHSE_ECOLI*
accession n°: P00547
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 931.448 (0), 956.34 (0), 962.402 (0), 1055.385 (0),
1112.426 (0), 1129.475 (0), 1159.503 (0), 1201.54 (0), 1217.522 (0),
1339.489 (0), 1374.558 (0), 1396.499 (0), 1431.558 (0), 1720.655 (0),
1777.661 (0) }
================
*FABI_ECOLI*
accession n: P0AEK4
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 997.838 (0), 1447.162 (0), 1486.1 (0), 1656.257 (0),
2009.576 (0), 2797.999 (0) }
Spot: 2D-001LCK (ecoli5-6)
pI: 5.33 Mw: 37266
%vol: 0.194923 %od: 0.386849
================
*KHSE_ECOLI*
accession n°: P00547
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 931.448 (0), 956.34 (0), 962.402 (0), 1055.385 (0),
1112.426 (0), 1129.475 (0), 1159.503 (0), 1201.54 (0), 1217.522 (0),
1339.489 (0), 1374.558 (0), 1396.499 (0), 1431.558 (0), 1720.655 (0),
1777.661 (0) }
================
*FABI_ECOLI*
accession n: P0AEK4
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 997.838 (0), 1447.162 (0), 1486.1 (0), 1656.257 (0),
2009.576 (0), 2797.999 (0) }
[Click to access protein entry]
Spot: 2D-001LF6 (ecoli5-6)
pI: 5.65 Mw: 34051
%vol: 0.185694 %od: 0.226817
================
*RTCB_ECOLI*
accession n°: P46850
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1365.676 (0), 1493.758 (0), 1507.752 (0), 1638.874 (0)
}
Spot: 2D-001LF6 (ecoli5-6)
pI: 5.65 Mw: 34051
%vol: 0.185694 %od: 0.226817
================
*RTCB_ECOLI*
accession n°: P46850
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1365.676 (0), 1493.758 (0), 1507.752 (0), 1638.874 (0)
}
[Click to access protein entry]
Spot: 2D-001L9E (ecoli5-6)
pI: 5.34 Mw: 42978
%vol: 0.198459 %od: 0.257817
================
*NADE_ECOLI*
accession n°: P18843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 911.627 (0), 1066.691 (0), 1114.768 (0), 1190.771 (0),
1235.791 (0), 1259.837 (0), 1263.79 (0), 1440.875 (0), 1521.011 (0),
1577.059 (0), 1603.077 (0), 1627.075 (0), 1639.107 (0), 1677.138 (0),
1795.225 (0), 2649.579 (0), 3735.304 (0) }
Spot: 2D-001L9E (ecoli5-6)
pI: 5.34 Mw: 42978
%vol: 0.198459 %od: 0.257817
================
*NADE_ECOLI*
accession n°: P18843
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 911.627 (0), 1066.691 (0), 1114.768 (0), 1190.771 (0),
1235.791 (0), 1259.837 (0), 1263.79 (0), 1440.875 (0), 1521.011 (0),
1577.059 (0), 1603.077 (0), 1627.075 (0), 1639.107 (0), 1677.138 (0),
1795.225 (0), 2649.579 (0), 3735.304 (0) }
[Click to access protein entry]
Spot: 2D-001LHT (ecoli5-6)
pI: 5.33 Mw: 31023
%vol: 0.133945 %od: 0.326431
================
*DAPB_ECOLI*
accession n°: P04036
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 870.525 (0), 954.583 (0), 970.57 (0), 1117.811 (0), 1619.03 (0),
1889.181 (0), 1899.158 (0), 1905.159 (0), 1915.165 (0), 2064.482 (0)
}
Spot: 2D-001LHT (ecoli5-6)
pI: 5.33 Mw: 31023
%vol: 0.133945 %od: 0.326431
================
*DAPB_ECOLI*
accession n°: P04036
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 870.525 (0), 954.583 (0), 970.57 (0), 1117.811 (0), 1619.03 (0),
1889.181 (0), 1899.158 (0), 1905.159 (0), 1915.165 (0), 2064.482 (0)
}
[Click to access protein entry]
Spot: 2D-001L1Y (ecoli5-6)
pI: 5.20 Mw: 51059
%vol: 0.17957 %od: 0.458662
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.599 (0), 1233.617 (0), 1796.913 (0), 1962.031 (0),
1964.982 (0), 2117.176 (0) }
Spot: 2D-001L1Y (ecoli5-6)
pI: 5.20 Mw: 51059
%vol: 0.17957 %od: 0.458662
================
*EFTU2_ECOLI*
accession n°: P0CE48
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1027.599 (0), 1233.617 (0), 1796.913 (0), 1962.031 (0),
1964.982 (0), 2117.176 (0) }
SWISS-2DPAGE Viewer
|
Back to the |
-Get more information by dragging your mouse pointer over any highlighted spot -Click on a highlighted spot to access all its associated protein entries
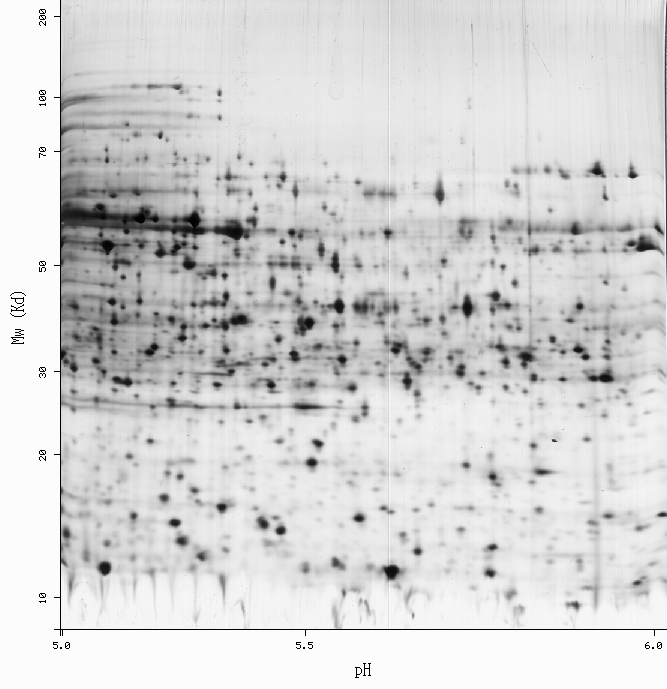
Back to the