Attention: World-2DPAGE is no longer maintained.
[Click to access protein entry]
Spot: 2D-001TNX (ecoli6-11)
pI: 7.13 Mw: 29388
%vol: 0.249316 %od: 0.540396
================
*GLNH_ECOLI*
accession n°: P0AEQ3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 984.259 (0), 1178.313 (0), 1306.426 (0), 1358.379 (0),
1567.498 (0), 1600.58 (0), 1683.601 (0), 1793.585 (0), 1829.647 (0),
1882.497 (0), 1898.508 (0), 2278.746 (0), 2294.766 (0)
}
Spot: 2D-001TNX (ecoli6-11)
pI: 7.13 Mw: 29388
%vol: 0.249316 %od: 0.540396
================
*GLNH_ECOLI*
accession n°: P0AEQ3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 984.259 (0), 1178.313 (0), 1306.426 (0), 1358.379 (0),
1567.498 (0), 1600.58 (0), 1683.601 (0), 1793.585 (0), 1829.647 (0),
1882.497 (0), 1898.508 (0), 2278.746 (0), 2294.766 (0)
}
[Click to access protein entry]
Spot: 2D-001TNE (ecoli6-11)
pI: 7.32 Mw: 30106
%vol: 0.20203 %od: 0.226082
================
*GLNH_ECOLI*
accession n°: P0AEQ3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1178.688 (0), 1601.085 (0), 1684.111 (0), 1794.128 (0),
1830.214 (0), 1883.131 (0), 1899.11 (0) }
Spot: 2D-001TNE (ecoli6-11)
pI: 7.32 Mw: 30106
%vol: 0.20203 %od: 0.226082
================
*GLNH_ECOLI*
accession n°: P0AEQ3
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1178.688 (0), 1601.085 (0), 1684.111 (0), 1794.128 (0),
1830.214 (0), 1883.131 (0), 1899.11 (0) }
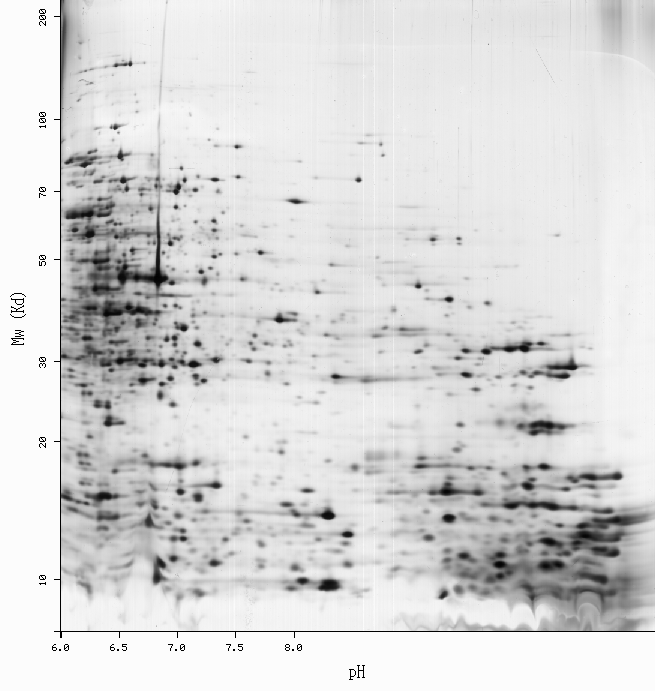
SWISS-2DPAGE Viewer
|
Back to the |
-Get more information by dragging your mouse pointer over any highlighted spot -Click on a highlighted spot to access all its associated protein entries
[estimated location]
Back to the