Attention: World-2DPAGE is no longer maintained.
[Click to access protein entry]
Spot: 2D-001TXO (ecoli6-11)
pI: 9.31 Mw: 13566
%vol: 0.254273 %od: 0.685578
================
*PLIG_ECOLI*
accession n°: P76002
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 877.577 (0), 992.688 (0), 1120.821 (0), 1263.874 (0),
1391.962 (0), 1506.944 (0), 1936.233 (0), 2863.988 (0)
}
Spot: 2D-001TXO (ecoli6-11)
pI: 9.31 Mw: 13566
%vol: 0.254273 %od: 0.685578
================
*PLIG_ECOLI*
accession n°: P76002
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 877.577 (0), 992.688 (0), 1120.821 (0), 1263.874 (0),
1391.962 (0), 1506.944 (0), 1936.233 (0), 2863.988 (0)
}
[Click to access protein entry]
Spot: 2D-001TZR (ecoli6-11)
pI: 9.14 Mw: 12279
%vol: 0.129962 %od: 0.242739
================
*PLIG_ECOLI*
accession n°: P76002
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 877.4 (0), 1263.628 (0), 1391.767 (0), 1506.64 (0),
1935.772 (0), 1936.037 (0), 2863.344 (0) }
Spot: 2D-001TZR (ecoli6-11)
pI: 9.14 Mw: 12279
%vol: 0.129962 %od: 0.242739
================
*PLIG_ECOLI*
accession n°: P76002
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 877.4 (0), 1263.628 (0), 1391.767 (0), 1506.64 (0),
1935.772 (0), 1936.037 (0), 2863.344 (0) }
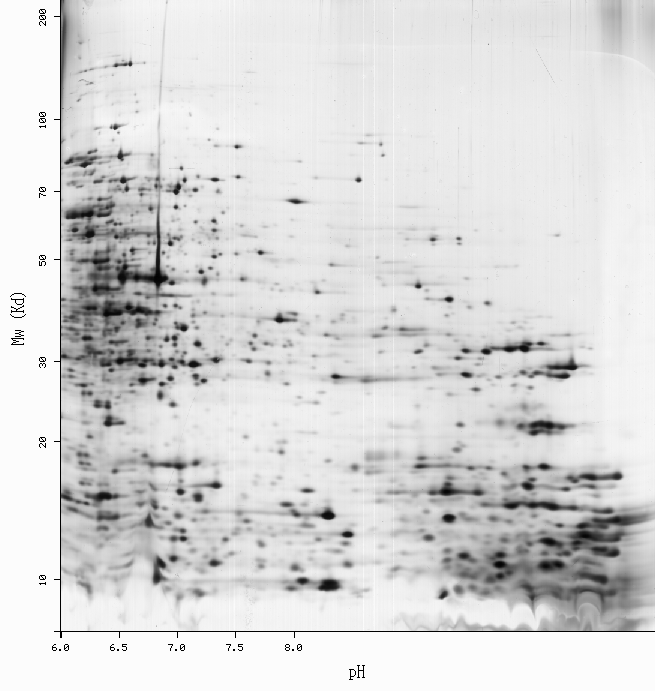
SWISS-2DPAGE Viewer
|
Back to the |
-Get more information by dragging your mouse pointer over any highlighted spot -Click on a highlighted spot to access all its associated protein entries
[estimated location]
Back to the