Attention: World-2DPAGE is no longer maintained.
[Click to access protein entry]
Spot: 2D-0014CA (liver_mouse)
pI: 5.29 Mw: 24557
%vol: 0.0325461 %od: 0.0367412
================
*CATB_MOUSE*
accession n°: P10605
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014CA (liver_mouse)
pI: 5.29 Mw: 24557
%vol: 0.0325461 %od: 0.0367412
================
*CATB_MOUSE*
accession n°: P10605
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0014BH (liver_mouse)
pI: 7.41 Mw: 24918
%vol: 0.110623 %od: 0.174339
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0014BH (liver_mouse)
pI: 7.41 Mw: 24918
%vol: 0.110623 %od: 0.174339
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-00135C (liver_mouse)
pI: 5.21 Mw: 65510
%vol: 0.0365718 %od: 0.0201164
================
*LMNB1_MOUSE*
accession n°: P14733
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-00135C (liver_mouse)
pI: 5.21 Mw: 65510
%vol: 0.0365718 %od: 0.0201164
================
*LMNB1_MOUSE*
accession n°: P14733
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0014LQ (liver_mouse)
pI: 8.16 Mw: 12126
%vol: 0.147308 %od: 0.703338
================
*HBA_MOUSE*
accession n°: P01942
Identification Methods:
* MAPPING: {Mi,PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1045.56 (0), 1529.759 (0), 1819.897 (0), 3050.449 (0)
}
Spot: 2D-0014LQ (liver_mouse)
pI: 8.16 Mw: 12126
%vol: 0.147308 %od: 0.703338
================
*HBA_MOUSE*
accession n°: P01942
Identification Methods:
* MAPPING: {Mi,PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1045.56 (0), 1529.759 (0), 1819.897 (0), 3050.449 (0)
}
[Click to access protein entry]
Spot: 2D-0013N7 (liver_mouse)
pI: 7.87 Mw: 42826
%vol: 0.147421 %od: 0.11342
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013N7 (liver_mouse)
pI: 7.87 Mw: 42826
%vol: 0.147421 %od: 0.11342
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013G5 (liver_mouse)
pI: 6.34 Mw: 49908
%vol: 0.13421 %od: 0.172881
================
*ALDH2_MOUSE*
accession n°: P47738
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013G5 (liver_mouse)
pI: 6.34 Mw: 49908
%vol: 0.13421 %od: 0.172881
================
*ALDH2_MOUSE*
accession n°: P47738
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013RC (liver_mouse)
pI: 6.59 Mw: 39289
%vol: 0.146344 %od: 0.200831
================
*IDHC_MOUSE*
accession n°: O88844
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 914.552 (0), 950.509 (0), 976.557 (0), 1009.447 (0),
1064.552 (0), 1087.568 (0), 1154.514 (0), 1170.573 (0), 1341.661 (0),
1398.691 (0), 1451.903 (0), 1525.671 (0), 1813.838 (0), 1864.922 (0),
2372.252 (0), 2626.066 (0) }
Spot: 2D-0013RC (liver_mouse)
pI: 6.59 Mw: 39289
%vol: 0.146344 %od: 0.200831
================
*IDHC_MOUSE*
accession n°: O88844
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 914.552 (0), 950.509 (0), 976.557 (0), 1009.447 (0),
1064.552 (0), 1087.568 (0), 1154.514 (0), 1170.573 (0), 1341.661 (0),
1398.691 (0), 1451.903 (0), 1525.671 (0), 1813.838 (0), 1864.922 (0),
2372.252 (0), 2626.066 (0) }
[Click to access protein entry]
Spot: 2D-0013A6 (liver_mouse)
pI: 5.29 Mw: 58144
%vol: 0.117937 %od: 0.132102
================
*CH60_MOUSE*
accession n°: P63038
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 855.452 (0), 912.58 (0), 960.512 (0), 1215.658 (0),
1389.748 (0), 1520.75 (0), 1601.788 (0), 1684.893 (0), 1905.069 (0),
2040.018 (0), 2365.296 (0), 2560.212 (0) }
Spot: 2D-0013A6 (liver_mouse)
pI: 5.29 Mw: 58144
%vol: 0.117937 %od: 0.132102
================
*CH60_MOUSE*
accession n°: P63038
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 855.452 (0), 912.58 (0), 960.512 (0), 1215.658 (0),
1389.748 (0), 1520.75 (0), 1601.788 (0), 1684.893 (0), 1905.069 (0),
2040.018 (0), 2365.296 (0), 2560.212 (0) }
[Click to access protein entry]
Spot: 2D-001335 (liver_mouse)
pI: 5.34 Mw: 68690
%vol: 0.127123 %od: 0.265855
================
*GRP75_MOUSE*
accession n°: P38647
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1242.721 (0), 1290.728 (0), 1462.771 (0), 1592.966 (0),
1694.889 (0), 2055.981 (0) }
Spot: 2D-001335 (liver_mouse)
pI: 5.34 Mw: 68690
%vol: 0.127123 %od: 0.265855
================
*GRP75_MOUSE*
accession n°: P38647
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1242.721 (0), 1290.728 (0), 1462.771 (0), 1592.966 (0),
1694.889 (0), 2055.981 (0) }
[Click to access protein entry]
Spot: 2D-0013LJ (liver_mouse)
pI: 6.50 Mw: 44527
%vol: 0.0405408 %od: 0.0451176
================
*GLNA_MOUSE*
accession n°: P15105
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 824.42 (0), 853.432 (0), 928.504 (0), 1002.487 (0),
1114.538 (0), 1146.53 (0), 1162.541 (0), 1794.853 (0), 1810.887 (0),
1814.864 (0), 1861.924 (0), 1918.937 (0), 2720.253 (0)
}
Spot: 2D-0013LJ (liver_mouse)
pI: 6.50 Mw: 44527
%vol: 0.0405408 %od: 0.0451176
================
*GLNA_MOUSE*
accession n°: P15105
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 824.42 (0), 853.432 (0), 928.504 (0), 1002.487 (0),
1114.538 (0), 1146.53 (0), 1162.541 (0), 1794.853 (0), 1810.887 (0),
1814.864 (0), 1861.924 (0), 1918.937 (0), 2720.253 (0)
}
[Click to access protein entry]
Spot: 2D-0012T1 (liver_mouse)
pI: 6.75 Mw: 99501
%vol: 0.117597 %od: 0.14193
================
*ACOC_MOUSE*
accession n°: P28271
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1037.538 (0), 1050.466 (0), 1100.557 (0), 1170.648 (0),
1458.815 (0), 1599.799 (0), 1815.977 (0), 1852.938 (0), 2060.148 (0),
2065.102 (0), 2090.153 (0), 2381.169 (0) }
Spot: 2D-0012T1 (liver_mouse)
pI: 6.75 Mw: 99501
%vol: 0.117597 %od: 0.14193
================
*ACOC_MOUSE*
accession n°: P28271
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1037.538 (0), 1050.466 (0), 1100.557 (0), 1170.648 (0),
1458.815 (0), 1599.799 (0), 1815.977 (0), 1852.938 (0), 2060.148 (0),
2065.102 (0), 2090.153 (0), 2381.169 (0) }
[Click to access protein entry]
Spot: 2D-00148I (liver_mouse)
pI: 8.22 Mw: 27086
%vol: 0.11238 %od: 0.0555549
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-00148I (liver_mouse)
pI: 8.22 Mw: 27086
%vol: 0.11238 %od: 0.0555549
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0012UI (liver_mouse)
pI: 4.88 Mw: 89679
%vol: 0.121169 %od: 0.0822534
================
*ENPL_MOUSE*
accession n°: P08113
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0012UI (liver_mouse)
pI: 4.88 Mw: 89679
%vol: 0.121169 %od: 0.0822534
================
*ENPL_MOUSE*
accession n°: P08113
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-001471 (liver_mouse)
pI: 6.74 Mw: 27889
%vol: 0.140334 %od: 0.32756
================
*CAH3_MOUSE*
accession n°: P16015
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-001471 (liver_mouse)
pI: 6.74 Mw: 27889
%vol: 0.140334 %od: 0.32756
================
*CAH3_MOUSE*
accession n°: P16015
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013BW (liver_mouse)
pI: 4.88 Mw: 55474
%vol: 0.141808 %od: 0.113717
================
*PDIA1_MOUSE*
accession n°: P09103
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 910.428 (0), 928.495 (0), 966.543 (0), 970.503 (0),
1066.506 (0), 1202.568 (0), 1216.586 (0), 1666.843 (0), 1780.83 (0),
1833.923 (0), 1965.082 (0) }
Spot: 2D-0013BW (liver_mouse)
pI: 4.88 Mw: 55474
%vol: 0.141808 %od: 0.113717
================
*PDIA1_MOUSE*
accession n°: P09103
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 910.428 (0), 928.495 (0), 966.543 (0), 970.503 (0),
1066.506 (0), 1202.568 (0), 1216.586 (0), 1666.843 (0), 1780.83 (0),
1833.923 (0), 1965.082 (0) }
[Click to access protein entry]
Spot: 2D-00138C (liver_mouse)
pI: 5.16 Mw: 59607
%vol: 0.0295409 %od: 0.0346855
================
*VIME_MOUSE*
accession n°: P20152
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 905.471 (0), 925.473 (0), 1093.542 (0), 1270.62 (0),
1296.637 (0), 1309.646 (0), 1320.651 (0), 1433.739 (0), 1444.696 (0),
1495.758 (0), 1533.809 (0), 1557.902 (0), 1838.892 (0), 2200.937 (0),
2216.903 (0) }
Spot: 2D-00138C (liver_mouse)
pI: 5.16 Mw: 59607
%vol: 0.0295409 %od: 0.0346855
================
*VIME_MOUSE*
accession n°: P20152
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 905.471 (0), 925.473 (0), 1093.542 (0), 1270.62 (0),
1296.637 (0), 1309.646 (0), 1320.651 (0), 1433.739 (0), 1444.696 (0),
1495.758 (0), 1533.809 (0), 1557.902 (0), 1838.892 (0), 2200.937 (0),
2216.903 (0) }
[Click to access protein entry]
Spot: 2D-0014IM (liver_mouse)
pI: 7.01 Mw: 14967
%vol: 0.0302214 %od: 0.0451227
================
*PPIA_MOUSE*
accession n°: P17742
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014IM (liver_mouse)
pI: 7.01 Mw: 14967
%vol: 0.0302214 %od: 0.0451227
================
*PPIA_MOUSE*
accession n°: P17742
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0013P6 (liver_mouse)
pI: 4.85 Mw: 40894
%vol: 0.0989424 %od: 0.110568
================
*RSSA_MOUSE*
accession n°: P14206
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013P6 (liver_mouse)
pI: 4.85 Mw: 40894
%vol: 0.0989424 %od: 0.110568
================
*RSSA_MOUSE*
accession n°: P14206
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013BE (liver_mouse)
pI: 5.71 Mw: 56408
%vol: 0.0891332 %od: 0.0769706
================
*PDIA3_MOUSE*
accession n°: P27773
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 877.493 (0), 995.544 (0), 997.506 (0), 1179.575 (0),
1188.519 (0), 1191.585 (0), 1258.656 (0), 1394.655 (0), 1397.698 (0),
2302.142 (0) }
Spot: 2D-0013BE (liver_mouse)
pI: 5.71 Mw: 56408
%vol: 0.0891332 %od: 0.0769706
================
*PDIA3_MOUSE*
accession n°: P27773
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 877.493 (0), 995.544 (0), 997.506 (0), 1179.575 (0),
1188.519 (0), 1191.585 (0), 1258.656 (0), 1394.655 (0), 1397.698 (0),
2302.142 (0) }
[Click to access protein entry]
Spot: 2D-0013GL (liver_mouse)
pI: 5.01 Mw: 49493
%vol: 0.131375 %od: 0.136963
================
*ATPB_MOUSE*
accession n°: P56480
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1435.879 (0), 1439.92 (0), 1651.06 (0), 1797.089 (0),
1843.055 (0), 1859.064 (0), 1919.258 (0), 1988.188 (0), 2298.211 (0)
}
Spot: 2D-0013GL (liver_mouse)
pI: 5.01 Mw: 49493
%vol: 0.131375 %od: 0.136963
================
*ATPB_MOUSE*
accession n°: P56480
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1435.879 (0), 1439.92 (0), 1651.06 (0), 1797.089 (0),
1843.055 (0), 1859.064 (0), 1919.258 (0), 1988.188 (0), 2298.211 (0)
}
[Click to access protein entry]
Spot: 2D-0012OD (liver_mouse)
pI: 5.96 Mw: 142608
%vol: 0.0987156 %od: 0.0600265
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012OD (liver_mouse)
pI: 5.96 Mw: 142608
%vol: 0.0987156 %od: 0.0600265
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0013SP (liver_mouse)
pI: 6.49 Mw: 38339
%vol: 0.152978 %od: 0.440894
================
*ARGI1_MOUSE*
accession n°: Q61176
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 911.506 (0), 948.644 (0), 1152.673 (0), 1444.851 (0),
1460.843 (0), 1853.105 (0), 1874.105 (0), 1910.136 (0), 1924.12 (0),
2018.249 (0), 2203.226 (0) }
Spot: 2D-0013SP (liver_mouse)
pI: 6.49 Mw: 38339
%vol: 0.152978 %od: 0.440894
================
*ARGI1_MOUSE*
accession n°: Q61176
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 911.506 (0), 948.644 (0), 1152.673 (0), 1444.851 (0),
1460.843 (0), 1853.105 (0), 1874.105 (0), 1910.136 (0), 1924.12 (0),
2018.249 (0), 2203.226 (0) }
[Click to access protein entry]
Spot: 2D-0012UM (liver_mouse)
pI: 5.53 Mw: 90000
%vol: 0.0227936 %od: 0.0202991
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012UM (liver_mouse)
pI: 5.53 Mw: 90000
%vol: 0.0227936 %od: 0.0202991
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0014K6 (liver_mouse)
pI: 4.48 Mw: 13430
%vol: 0.0249482 %od: 0.0360309
================
*ATPD_MOUSE*
accession n°: P99033
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014K6 (liver_mouse)
pI: 4.48 Mw: 13430
%vol: 0.0249482 %od: 0.0360309
================
*ATPD_MOUSE*
accession n°: P99033
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0014I5 (liver_mouse)
pI: 6.98 Mw: 15443
%vol: 0.0314687 %od: 0.0389027
================
*NDKB_MOUSE*
accession n°: Q01768
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 994.482 (0), 1051.499 (0), 1175.666 (0), 1344.76 (0),
1801.919 (0), 1960.021 (0), 2125.064 (0) }
Spot: 2D-0014I5 (liver_mouse)
pI: 6.98 Mw: 15443
%vol: 0.0314687 %od: 0.0389027
================
*NDKB_MOUSE*
accession n°: Q01768
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 994.482 (0), 1051.499 (0), 1175.666 (0), 1344.76 (0),
1801.919 (0), 1960.021 (0), 2125.064 (0) }
[Click to access protein entry]
Spot: 2D-0013N6 (liver_mouse)
pI: 7.74 Mw: 42826
%vol: 0.139483 %od: 0.0502005
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013N6 (liver_mouse)
pI: 7.74 Mw: 42826
%vol: 0.139483 %od: 0.0502005
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0014GL (liver_mouse)
pI: 4.86 Mw: 18512
%vol: 0.144983 %od: 0.386753
================
*MUP2_MOUSE*
accession n°: P11589
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.453 (0), 1126.596 (0), 1258.641 (0), 1368.79 (0),
1486.75 (0), 1502.691 (0), 1596.861 (0), 1839.863 (0), 2009.927 (0)
}
Spot: 2D-0014GL (liver_mouse)
pI: 4.86 Mw: 18512
%vol: 0.144983 %od: 0.386753
================
*MUP2_MOUSE*
accession n°: P11589
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.453 (0), 1126.596 (0), 1258.641 (0), 1368.79 (0),
1486.75 (0), 1502.691 (0), 1596.861 (0), 1839.863 (0), 2009.927 (0)
}
[Click to access protein entry]
Spot: 2D-0014GD (liver_mouse)
pI: 4.88 Mw: 19046
%vol: 0.145834 %od: 0.304259
================
*MUP6_MOUSE*
accession n°: P02762
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1415.713 (0), 1486.679 (0), 1502.729 (0), 1596.799 (0),
1839.865 (0), 2009.971 (0) }
================
*MUP2_MOUSE*
accession n: P11589
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1368.787 (0), 1486.679 (0), 1502.729 (0), 1596.799 (0),
1839.865 (0), 2009.971 (0) }
Spot: 2D-0014GD (liver_mouse)
pI: 4.88 Mw: 19046
%vol: 0.145834 %od: 0.304259
================
*MUP6_MOUSE*
accession n°: P02762
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1415.713 (0), 1486.679 (0), 1502.729 (0), 1596.799 (0),
1839.865 (0), 2009.971 (0) }
================
*MUP2_MOUSE*
accession n: P11589
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1368.787 (0), 1486.679 (0), 1502.729 (0), 1596.799 (0),
1839.865 (0), 2009.971 (0) }
[Click to access protein entry]
Spot: 2D-0013NW (liver_mouse)
pI: 6.50 Mw: 42000
%vol: 0.0442264 %od: 0.045565
================
*IDHC_MOUSE*
accession n°: O88844
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 914.529 (0), 950.493 (0), 976.557 (0), 1009.456 (0),
1064.562 (0), 1154.554 (0), 1250.621 (0), 1341.672 (0), 1398.685 (0),
1451.898 (0), 1525.689 (0), 1768.82 (0), 1813.845 (0), 1864.902 (0),
2372.232 (0) }
Spot: 2D-0013NW (liver_mouse)
pI: 6.50 Mw: 42000
%vol: 0.0442264 %od: 0.045565
================
*IDHC_MOUSE*
accession n°: O88844
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 914.529 (0), 950.493 (0), 976.557 (0), 1009.456 (0),
1064.562 (0), 1154.554 (0), 1250.621 (0), 1341.672 (0), 1398.685 (0),
1451.898 (0), 1525.689 (0), 1768.82 (0), 1813.845 (0), 1864.902 (0),
2372.232 (0) }
[Click to access protein entry]
Spot: 2D-001HE9 (liver_mouse)
pI: 4.23 Mw: 13799
%vol: 0.00549995 %od: 0.0103792
================
*CALM_MOUSE*
accession n°: P62204
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 956.416 (0), 1093.477 (0), 1265.618 (0), 1652.923 (0),
1754.886 (0) }
Spot: 2D-001HE9 (liver_mouse)
pI: 4.23 Mw: 13799
%vol: 0.00549995 %od: 0.0103792
================
*CALM_MOUSE*
accession n°: P62204
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 956.416 (0), 1093.477 (0), 1265.618 (0), 1652.923 (0),
1754.886 (0) }
[Click to access protein entry]
Spot: 2D-0012PQ (liver_mouse)
pI: 6.16 Mw: 121389
%vol: 0.104783 %od: 0.0847172
================
*PYC_MOUSE*
accession n°: Q05920
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012PQ (liver_mouse)
pI: 6.16 Mw: 121389
%vol: 0.104783 %od: 0.0847172
================
*PYC_MOUSE*
accession n°: Q05920
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0013LF (liver_mouse)
pI: 5.15 Mw: 44157
%vol: 0.049386 %od: 0.0529955
================
*ACTB_MOUSE*
accession n°: P99041
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1132.456 (0), 1198.649 (0), 1516.718 (0), 1790.835 (0)
}
Spot: 2D-0013LF (liver_mouse)
pI: 5.15 Mw: 44157
%vol: 0.049386 %od: 0.0529955
================
*ACTB_MOUSE*
accession n°: P99041
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1132.456 (0), 1198.649 (0), 1516.718 (0), 1790.835 (0)
}
[Click to access protein entry]
Spot: 2D-0013NO (liver_mouse)
pI: 5.26 Mw: 41907
%vol: 0.138973 %od: 0.317681
================
*ACTB_MOUSE*
accession n°: P99041
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013NO (liver_mouse)
pI: 5.26 Mw: 41907
%vol: 0.138973 %od: 0.317681
================
*ACTB_MOUSE*
accession n°: P99041
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0014KV (liver_mouse)
pI: 5.97 Mw: 12748
%vol: 0.0588551 %od: 0.0946948
================
*COX5B_MOUSE*
accession n°: P19536
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014KV (liver_mouse)
pI: 5.97 Mw: 12748
%vol: 0.0588551 %od: 0.0946948
================
*COX5B_MOUSE*
accession n°: P19536
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0013E5 (liver_mouse)
pI: 5.80 Mw: 52618
%vol: 0.137896 %od: 0.438767
================
*SBP1_MOUSE*
accession n°: P17563
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013E5 (liver_mouse)
pI: 5.80 Mw: 52618
%vol: 0.137896 %od: 0.438767
================
*SBP1_MOUSE*
accession n°: P17563
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013YH (liver_mouse)
pI: 5.03 Mw: 33718
%vol: 0.0347008 %od: 0.034928
================
*RGN_MOUSE*
accession n°: Q64374
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 960.45 (0), 1057.562 (0), 1138.572 (0), 1195.601 (0),
1232.628 (0), 1283.758 (0), 1394.568 (0), 1410.6 (0), 1714.888 (0),
1856.02 (0), 1871.913 (0), 2152.033 (0) }
Spot: 2D-0013YH (liver_mouse)
pI: 5.03 Mw: 33718
%vol: 0.0347008 %od: 0.034928
================
*RGN_MOUSE*
accession n°: Q64374
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 960.45 (0), 1057.562 (0), 1138.572 (0), 1195.601 (0),
1232.628 (0), 1283.758 (0), 1394.568 (0), 1410.6 (0), 1714.888 (0),
1856.02 (0), 1871.913 (0), 2152.033 (0) }
[Click to access protein entry]
Spot: 2D-0013EL (liver_mouse)
pI: 7.65 Mw: 52472
%vol: 0.12202 %od: 0.200317
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013EL (liver_mouse)
pI: 7.65 Mw: 52472
%vol: 0.12202 %od: 0.200317
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0012UQ (liver_mouse)
pI: 5.69 Mw: 89359
%vol: 0.107504 %od: 0.0941933
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012UQ (liver_mouse)
pI: 5.69 Mw: 89359
%vol: 0.107504 %od: 0.0941933
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0012PT (liver_mouse)
pI: 6.10 Mw: 121389
%vol: 0.0527881 %od: 0.0190237
================
*PYC_MOUSE*
accession n°: Q05920
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012PT (liver_mouse)
pI: 6.10 Mw: 121389
%vol: 0.0527881 %od: 0.0190237
================
*PYC_MOUSE*
accession n°: Q05920
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0013IV (liver_mouse)
pI: 6.16 Mw: 47076
%vol: 0.131432 %od: 0.11239
================
*ENOA_MOUSE*
accession n°: P17182
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013IV (liver_mouse)
pI: 6.16 Mw: 47076
%vol: 0.131432 %od: 0.11239
================
*ENOA_MOUSE*
accession n°: P17182
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013F0 (liver_mouse)
pI: 6.47 Mw: 52035
%vol: 0.0974682 %od: 0.125602
================
*DHE3_MOUSE*
accession n°: P26443
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.576 (0), 1016.457 (0), 1059.559 (0), 1196.685 (0),
1425.668 (0), 1723.897 (0), 1764.868 (0), 1894.112 (0), 1931.961 (0),
1936.899 (0) }
Spot: 2D-0013F0 (liver_mouse)
pI: 6.47 Mw: 52035
%vol: 0.0974682 %od: 0.125602
================
*DHE3_MOUSE*
accession n°: P26443
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.576 (0), 1016.457 (0), 1059.559 (0), 1196.685 (0),
1425.668 (0), 1723.897 (0), 1764.868 (0), 1894.112 (0), 1931.961 (0),
1936.899 (0) }
[Click to access protein entry]
Spot: 2D-0014CV (liver_mouse)
pI: 4.81 Mw: 24050
%vol: 0.058685 %od: 0.0767573
================
*TCTP_MOUSE*
accession n°: P63028
Identification Methods:
* MAPPING: {Mi,Gm}
Spot: 2D-0014CV (liver_mouse)
pI: 4.81 Mw: 24050
%vol: 0.058685 %od: 0.0767573
================
*TCTP_MOUSE*
accession n°: P63028
Identification Methods:
* MAPPING: {Mi,Gm}
[Click to access protein entry]
Spot: 2D-0014NN (liver_mouse)
pI: 5.31 Mw: 9788
%vol: 0.0286337 %od: 0.052823
================
*ATP5J_MOUSE*
accession n°: P97450
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014NN (liver_mouse)
pI: 5.31 Mw: 9788
%vol: 0.0286337 %od: 0.052823
================
*ATP5J_MOUSE*
accession n°: P97450
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0014KP (liver_mouse)
pI: 8.64 Mw: 12881
%vol: 0.0645252 %od: 0.147927
================
*FABPL_MOUSE*
accession n°: P12710
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 892.468 (0), 1054.514 (0), 1196.661 (0), 1804.835 (0),
2132.939 (0), 2401.303 (0) }
Spot: 2D-0014KP (liver_mouse)
pI: 8.64 Mw: 12881
%vol: 0.0645252 %od: 0.147927
================
*FABPL_MOUSE*
accession n°: P12710
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 892.468 (0), 1054.514 (0), 1196.661 (0), 1804.835 (0),
2132.939 (0), 2401.303 (0) }
[Click to access protein entry]
Spot: 2D-00138A (liver_mouse)
pI: 6.20 Mw: 60053
%vol: 0.0962775 %od: 0.0627139
================
*PGMU_MOUSE*
accession n°: P99030
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-00138A (liver_mouse)
pI: 6.20 Mw: 60053
%vol: 0.0962775 %od: 0.0627139
================
*PGMU_MOUSE*
accession n°: P99030
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0012V4 (liver_mouse)
pI: 6.90 Mw: 89040
%vol: 0.0801178 %od: 0.0411002
================
*ACOC_MOUSE*
accession n°: P28271
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1458.748 (0), 1599.776 (0), 1852.938 (0), 2060.13 (0),
2090.132 (0), 2381.167 (0) }
Spot: 2D-0012V4 (liver_mouse)
pI: 6.90 Mw: 89040
%vol: 0.0801178 %od: 0.0411002
================
*ACOC_MOUSE*
accession n°: P28271
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1458.748 (0), 1599.776 (0), 1852.938 (0), 2060.13 (0),
2090.132 (0), 2381.167 (0) }
[Click to access protein entry]
Spot: 2D-0014LE (liver_mouse)
pI: 4.91 Mw: 12433
%vol: 0.104442 %od: 0.223049
================
*THIO_MOUSE*
accession n°: P10639
Identification Methods:
* MAPPING: {Mi,PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1237.544 (0), 1308.585 (0), 1320.529 (0), 1365.532 (0),
1624.74 (0) }
Spot: 2D-0014LE (liver_mouse)
pI: 4.91 Mw: 12433
%vol: 0.104442 %od: 0.223049
================
*THIO_MOUSE*
accession n°: P10639
Identification Methods:
* MAPPING: {Mi,PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1237.544 (0), 1308.585 (0), 1320.529 (0), 1365.532 (0),
1624.74 (0) }
[Click to access protein entry]
Spot: 2D-0014IX (liver_mouse)
pI: 6.15 Mw: 14537
%vol: 0.131318 %od: 0.466741
================
*SODC_MOUSE*
accession n°: P08228
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0014IX (liver_mouse)
pI: 6.15 Mw: 14537
%vol: 0.131318 %od: 0.466741
================
*SODC_MOUSE*
accession n°: P08228
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0012NB (liver_mouse)
pI: 6.17 Mw: 160000
%vol: 0.107391 %od: 0.0604636
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012NB (liver_mouse)
pI: 6.17 Mw: 160000
%vol: 0.107391 %od: 0.0604636
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0014L5 (liver_mouse)
pI: 7.42 Mw: 12642
%vol: 0.0887363 %od: 0.301556
================
*HBB1_MOUSE*
accession n°: P02088
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1126.57 (0), 1274.739 (0), 1294.684 (0), 1756.919 (0)
}
Spot: 2D-0014L5 (liver_mouse)
pI: 7.42 Mw: 12642
%vol: 0.0887363 %od: 0.301556
================
*HBB1_MOUSE*
accession n°: P02088
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1126.57 (0), 1274.739 (0), 1294.684 (0), 1756.919 (0)
}
[Click to access protein entry]
Spot: 2D-0014CT (liver_mouse)
pI: 7.22 Mw: 24100
%vol: 0.124174 %od: 0.22426
================
*SODM_MOUSE*
accession n°: P09671
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014CT (liver_mouse)
pI: 7.22 Mw: 24100
%vol: 0.124174 %od: 0.22426
================
*SODM_MOUSE*
accession n°: P09671
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0013MW (liver_mouse)
pI: 7.53 Mw: 42826
%vol: 0.169875 %od: 0.467164
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013MW (liver_mouse)
pI: 7.53 Mw: 42826
%vol: 0.169875 %od: 0.467164
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0012ND (liver_mouse)
pI: 6.22 Mw: 160000
%vol: 0.125025 %od: 0.0977957
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1007.473 (0), 1162.595 (0), 1222.635 (0), 1298.677 (0),
1353.729 (0), 1408.745 (0), 1475.745 (0), 1484.879 (0), 1588.822 (0),
1637.918 (0), 1707.768 (0) }
Spot: 2D-0012ND (liver_mouse)
pI: 6.22 Mw: 160000
%vol: 0.125025 %od: 0.0977957
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1007.473 (0), 1162.595 (0), 1222.635 (0), 1298.677 (0),
1353.729 (0), 1408.745 (0), 1475.745 (0), 1484.879 (0), 1588.822 (0),
1637.918 (0), 1707.768 (0) }
[Click to access protein entry]
Spot: 2D-0012XD (liver_mouse)
pI: 5.02 Mw: 82305
%vol: 0.0667932 %od: 0.033405
================
*HS90B_MOUSE*
accession n°: P11499
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0012XD (liver_mouse)
pI: 5.02 Mw: 82305
%vol: 0.0667932 %od: 0.033405
================
*HS90B_MOUSE*
accession n°: P11499
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013Q1 (liver_mouse)
pI: 6.85 Mw: 40352
%vol: 0.08369 %od: 0.0544724
================
*AATC_MOUSE*
accession n°: P05201
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 848.416 (0), 948.493 (0), 997.36 (0), 1012.454 (0),
1055.574 (0), 1121.552 (0), 1271.672 (0), 1297.655 (0), 1403.733 (0),
1660.824 (0) }
Spot: 2D-0013Q1 (liver_mouse)
pI: 6.85 Mw: 40352
%vol: 0.08369 %od: 0.0544724
================
*AATC_MOUSE*
accession n°: P05201
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 848.416 (0), 948.493 (0), 997.36 (0), 1012.454 (0),
1055.574 (0), 1121.552 (0), 1271.672 (0), 1297.655 (0), 1403.733 (0),
1660.824 (0) }
[Click to access protein entry]
Spot: 2D-0013BV (liver_mouse)
pI: 4.82 Mw: 55784
%vol: 0.132963 %od: 0.0592001
================
*PDIA1_MOUSE*
accession n°: P09103
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 850.47 (0), 905.518 (0), 910.461 (0), 966.582 (0), 1066.542 (0),
1216.615 (0), 1666.87 (0), 1833.911 (0), 1965.099 (0)
}
Spot: 2D-0013BV (liver_mouse)
pI: 4.82 Mw: 55784
%vol: 0.132963 %od: 0.0592001
================
*PDIA1_MOUSE*
accession n°: P09103
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 850.47 (0), 905.518 (0), 910.461 (0), 966.582 (0), 1066.542 (0),
1216.615 (0), 1666.87 (0), 1833.911 (0), 1965.099 (0)
}
[Click to access protein entry]
Spot: 2D-001366 (liver_mouse)
pI: 5.29 Mw: 63270
%vol: 0.0205823 %od: 0.0166538
================
*ACADV_MOUSE*
accession n°: P50544
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 934.537 (0), 1184.573 (0), 1228.626 (0), 1316.728 (0),
1880.008 (0), 2017.893 (0), 2577.284 (0) }
Spot: 2D-001366 (liver_mouse)
pI: 5.29 Mw: 63270
%vol: 0.0205823 %od: 0.0166538
================
*ACADV_MOUSE*
accession n°: P50544
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 934.537 (0), 1184.573 (0), 1228.626 (0), 1316.728 (0),
1880.008 (0), 2017.893 (0), 2577.284 (0) }
[Click to access protein entry]
Spot: 2D-0013JB (liver_mouse)
pI: 5.26 Mw: 46554
%vol: 0.105406 %od: 0.12979
================
*K1C18_MOUSE*
accession n°: P05784
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013JB (liver_mouse)
pI: 5.26 Mw: 46554
%vol: 0.105406 %od: 0.12979
================
*K1C18_MOUSE*
accession n°: P05784
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-00134L (liver_mouse)
pI: 5.42 Mw: 66000
%vol: 0.0468345 %od: 0.0356212
================
*ALBU_MOUSE*
accession n°: P07724
Identification Methods:
* MAPPING: {Mi,Gm}
Spot: 2D-00134L (liver_mouse)
pI: 5.42 Mw: 66000
%vol: 0.0468345 %od: 0.0356212
================
*ALBU_MOUSE*
accession n°: P07724
Identification Methods:
* MAPPING: {Mi,Gm}
[Click to access protein entry]
Spot: 2D-0013IE (liver_mouse)
pI: 5.88 Mw: 47470
%vol: 0.0980919 %od: 0.0618107
================
*OAT_MOUSE*
accession n°: P29758
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 968.637 (0), 1225.628 (0), 1241.513 (0), 1281.615 (0),
1639.749 (0), 1736.847 (0), 1810.949 (0), 1952.927 (0), 2107.959 (0)
}
Spot: 2D-0013IE (liver_mouse)
pI: 5.88 Mw: 47470
%vol: 0.0980919 %od: 0.0618107
================
*OAT_MOUSE*
accession n°: P29758
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 968.637 (0), 1225.628 (0), 1241.513 (0), 1281.615 (0),
1639.749 (0), 1736.847 (0), 1810.949 (0), 1952.927 (0), 2107.959 (0)
}
[Click to access protein entry]
Spot: 2D-0014KE (liver_mouse)
pI: 9.00 Mw: 13016
%vol: 0.165225 %od: 1.4914
================
*FABPL_MOUSE*
accession n°: P12710
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 892.475 (0), 1054.516 (0), 1133.659 (0), 1182.624 (0),
1196.661 (0), 1246.726 (0), 1804.809 (0), 2132.891 (0), 2401.192 (0)
}
Spot: 2D-0014KE (liver_mouse)
pI: 9.00 Mw: 13016
%vol: 0.165225 %od: 1.4914
================
*FABPL_MOUSE*
accession n°: P12710
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 892.475 (0), 1054.516 (0), 1133.659 (0), 1182.624 (0),
1196.661 (0), 1246.726 (0), 1804.809 (0), 2132.891 (0), 2401.192 (0)
}
[Click to access protein entry]
Spot: 2D-0014M9 (liver_mouse)
pI: 4.99 Mw: 11731
%vol: 0.0337368 %od: 0.0345148
================
*ULAL_MOUSE*
accession n°: P99032
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014M9 (liver_mouse)
pI: 4.99 Mw: 11731
%vol: 0.0337368 %od: 0.0345148
================
*ULAL_MOUSE*
accession n°: P99032
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0014LH (liver_mouse)
pI: 5.06 Mw: 12381
%vol: 0.0913446 %od: 0.172195
================
*COX5A_MOUSE*
accession n°: P12787
Identification Methods:
* MAPPING: {Mi,PMF}
peptide masses: { (TRYPSIN)
$m/Z= 992.563 (0), 1148.645 (0), 1616.67 (0), 1648.806 (0),
2053.025 (0), 3329.648 (0), 3428.733 (0) }
Spot: 2D-0014LH (liver_mouse)
pI: 5.06 Mw: 12381
%vol: 0.0913446 %od: 0.172195
================
*COX5A_MOUSE*
accession n°: P12787
Identification Methods:
* MAPPING: {Mi,PMF}
peptide masses: { (TRYPSIN)
$m/Z= 992.563 (0), 1148.645 (0), 1616.67 (0), 1648.806 (0),
2053.025 (0), 3329.648 (0), 3428.733 (0) }
[Click to access protein entry]
Spot: 2D-0013XZ (liver_mouse)
pI: 5.08 Mw: 33858
%vol: 0.136421 %od: 0.482884
================
*RGN_MOUSE*
accession n°: Q64374
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 989.542 (0), 1057.611 (0), 1138.597 (0), 1195.618 (0),
1232.656 (0), 1264.643 (0), 1283.788 (0), 1353.616 (0), 1394.648 (0),
1410.632 (0), 1657.88 (0), 1714.947 (0), 1856.012 (0), 1871.977 (0),
2152.109 (0), 2280.092 (0), 2478.146 (0), 2659.376 (0)
}
Spot: 2D-0013XZ (liver_mouse)
pI: 5.08 Mw: 33858
%vol: 0.136421 %od: 0.482884
================
*RGN_MOUSE*
accession n°: Q64374
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 989.542 (0), 1057.611 (0), 1138.597 (0), 1195.618 (0),
1232.656 (0), 1264.643 (0), 1283.788 (0), 1353.616 (0), 1394.648 (0),
1410.632 (0), 1657.88 (0), 1714.947 (0), 1856.012 (0), 1871.977 (0),
2152.109 (0), 2280.092 (0), 2478.146 (0), 2659.376 (0)
}
[Click to access protein entry]
Spot: 2D-0013JA (liver_mouse)
pI: 5.21 Mw: 46684
%vol: 0.0735406 %od: 0.0825163
================
*K1C18_MOUSE*
accession n°: P05784
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013JA (liver_mouse)
pI: 5.21 Mw: 46684
%vol: 0.0735406 %od: 0.0825163
================
*K1C18_MOUSE*
accession n°: P05784
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0014CM (liver_mouse)
pI: 5.27 Mw: 24201
%vol: 0.0474015 %od: 0.0542555
================
*APOA1_MOUSE*
accession n°: Q00623
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0014CM (liver_mouse)
pI: 5.27 Mw: 24201
%vol: 0.0474015 %od: 0.0542555
================
*APOA1_MOUSE*
accession n°: Q00623
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0014IV (liver_mouse)
pI: 7.65 Mw: 14628
%vol: 0.0570974 %od: 0.0809211
================
*PRDX5_MOUSE*
accession n°: P99029
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014IV (liver_mouse)
pI: 7.65 Mw: 14628
%vol: 0.0570974 %od: 0.0809211
================
*PRDX5_MOUSE*
accession n°: P99029
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0012N3 (liver_mouse)
pI: 6.35 Mw: 160000
%vol: 0.14572 %od: 0.176654
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 822.499 (0), 1023.535 (0), 1090.675 (0), 1162.696 (0),
1221.774 (0), 1298.733 (0), 1353.787 (0), 1484.918 (0), 1588.853 (0),
1637.965 (0), 2553.323 (0) }
Spot: 2D-0012N3 (liver_mouse)
pI: 6.35 Mw: 160000
%vol: 0.14572 %od: 0.176654
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 822.499 (0), 1023.535 (0), 1090.675 (0), 1162.696 (0),
1221.774 (0), 1298.733 (0), 1353.787 (0), 1484.918 (0), 1588.853 (0),
1637.965 (0), 2553.323 (0) }
[Click to access protein entry]
Spot: 2D-00132H (liver_mouse)
pI: 4.99 Mw: 70356
%vol: 0.0551697 %od: 0.0300175
================
*GRP78_MOUSE*
accession n°: P20029
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 919.464 (0), 931.404 (0), 986.523 (0), 1074.534 (0),
1191.664 (0), 1228.649 (0), 1307.731 (0), 1323.746 (0), 1329.656 (0),
1430.716 (0), 1460.807 (0), 1528.796 (0), 1566.832 (0), 1588.901 (0),
1659.846 (0), 1773.931 (0), 1888.047 (0), 1934.1 (0), 1974.941 (0),
1999.175 (0), 2016.131 (0), 2149.08 (0) }
Spot: 2D-00132H (liver_mouse)
pI: 4.99 Mw: 70356
%vol: 0.0551697 %od: 0.0300175
================
*GRP78_MOUSE*
accession n°: P20029
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 919.464 (0), 931.404 (0), 986.523 (0), 1074.534 (0),
1191.664 (0), 1228.649 (0), 1307.731 (0), 1323.746 (0), 1329.656 (0),
1430.716 (0), 1460.807 (0), 1528.796 (0), 1566.832 (0), 1588.901 (0),
1659.846 (0), 1773.931 (0), 1888.047 (0), 1934.1 (0), 1974.941 (0),
1999.175 (0), 2016.131 (0), 2149.08 (0) }
[Click to access protein entry]
Spot: 2D-001393 (liver_mouse)
pI: 5.75 Mw: 59312
%vol: 0.126329 %od: 0.0711262
================
*TCPE_MOUSE*
accession n°: P80316
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-001393 (liver_mouse)
pI: 5.75 Mw: 59312
%vol: 0.126329 %od: 0.0711262
================
*TCPE_MOUSE*
accession n°: P80316
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-00139S (liver_mouse)
pI: 5.32 Mw: 58000
%vol: 0.141184 %od: 0.36955
================
*CH60_MOUSE*
accession n°: P63038
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 855.519 (0), 1006.528 (0), 1344.735 (0), 1389.726 (0),
1627.883 (0), 1684.92 (0), 1905.045 (0), 2040.013 (0), 2365.321 (0),
2560.215 (0) }
Spot: 2D-00139S (liver_mouse)
pI: 5.32 Mw: 58000
%vol: 0.141184 %od: 0.36955
================
*CH60_MOUSE*
accession n°: P63038
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 855.519 (0), 1006.528 (0), 1344.735 (0), 1389.726 (0),
1627.883 (0), 1684.92 (0), 1905.045 (0), 2040.013 (0), 2365.321 (0),
2560.215 (0) }
[Click to access protein entry]
Spot: 2D-0012UP (liver_mouse)
pI: 5.62 Mw: 90000
%vol: 0.0593655 %od: 0.034163
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012UP (liver_mouse)
pI: 5.62 Mw: 90000
%vol: 0.0593655 %od: 0.034163
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0014AY (liver_mouse)
pI: 8.63 Mw: 25390
%vol: 0.0412214 %od: 0.0533301
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0014AY (liver_mouse)
pI: 8.63 Mw: 25390
%vol: 0.0412214 %od: 0.0533301
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013OY (liver_mouse)
pI: 6.77 Mw: 40894
%vol: 0.163241 %od: 0.415586
================
*FAAA_MOUSE*
accession n°: P35505
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013OY (liver_mouse)
pI: 6.77 Mw: 40894
%vol: 0.163241 %od: 0.415586
================
*FAAA_MOUSE*
accession n°: P35505
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-00142R (liver_mouse)
pI: 5.69 Mw: 30569
%vol: 0.0429791 %od: 0.0593675
================
*KHK_MOUSE*
accession n°: P97328
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 910.56 (0), 1021.484 (0), 1056.635 (0), 1193.725 (0),
1662.926 (0), 1676.912 (0), 2021.039 (0), 2141.161 (0)
}
Spot: 2D-00142R (liver_mouse)
pI: 5.69 Mw: 30569
%vol: 0.0429791 %od: 0.0593675
================
*KHK_MOUSE*
accession n°: P97328
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 910.56 (0), 1021.484 (0), 1056.635 (0), 1193.725 (0),
1662.926 (0), 1676.912 (0), 2021.039 (0), 2141.161 (0)
}
[Click to access protein entry]
Spot: 2D-0012OI (liver_mouse)
pI: 5.91 Mw: 140977
%vol: 0.0491025 %od: 0.0278406
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012OI (liver_mouse)
pI: 5.91 Mw: 140977
%vol: 0.0491025 %od: 0.0278406
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-00149R (liver_mouse)
pI: 7.26 Mw: 26307
%vol: 0.0949734 %od: 0.153768
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 976.503 (0), 1015.572 (0), 1105.546 (0), 1264.612 (0),
1899.014 (0), 1993.991 (0) }
Spot: 2D-00149R (liver_mouse)
pI: 7.26 Mw: 26307
%vol: 0.0949734 %od: 0.153768
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 976.503 (0), 1015.572 (0), 1105.546 (0), 1264.612 (0),
1899.014 (0), 1993.991 (0) }
[Click to access protein entry]
Spot: 2D-0013AO (liver_mouse)
pI: 6.36 Mw: 57678
%vol: 0.0239277 %od: 0.00679197
================
*HYES_MOUSE*
accession n°: P34914
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013AO (liver_mouse)
pI: 6.36 Mw: 57678
%vol: 0.0239277 %od: 0.00679197
================
*HYES_MOUSE*
accession n°: P34914
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-00135F (liver_mouse)
pI: 6.93 Mw: 65347
%vol: 0.107958 %od: 0.125925
================
*TKT_MOUSE*
accession n°: P40142
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 936.396 (0), 1402.775 (0), 1413.771 (0), 1651.778 (0),
2020.084 (0), 2024.967 (0), 2481.212 (0), 3188.67 (0)
}
Spot: 2D-00135F (liver_mouse)
pI: 6.93 Mw: 65347
%vol: 0.107958 %od: 0.125925
================
*TKT_MOUSE*
accession n°: P40142
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 936.396 (0), 1402.775 (0), 1413.771 (0), 1651.778 (0),
2020.084 (0), 2024.967 (0), 2481.212 (0), 3188.67 (0)
}
[Click to access protein entry]
Spot: 2D-00149J (liver_mouse)
pI: 6.26 Mw: 26252
%vol: 0.143679 %od: 0.387014
================
*PRDX6_MOUSE*
accession n°: O08709
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 906.489 (0), 1021.585 (0), 1057.603 (0), 1149.629 (0),
1191.687 (0), 1270.704 (0), 1338.656 (0), 1395.671 (0), 2030.983 (0),
2142.067 (0) }
Spot: 2D-00149J (liver_mouse)
pI: 6.26 Mw: 26252
%vol: 0.143679 %od: 0.387014
================
*PRDX6_MOUSE*
accession n°: O08709
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 906.489 (0), 1021.585 (0), 1057.603 (0), 1149.629 (0),
1191.687 (0), 1270.704 (0), 1338.656 (0), 1395.671 (0), 2030.983 (0),
2142.067 (0) }
[Click to access protein entry]
Spot: 2D-0012UR (liver_mouse)
pI: 5.79 Mw: 90000
%vol: 0.0297677 %od: 0.0196059
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012UR (liver_mouse)
pI: 5.79 Mw: 90000
%vol: 0.0297677 %od: 0.0196059
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0014AG (liver_mouse)
pI: 6.88 Mw: 25710
%vol: 0.0987723 %od: 0.0764068
================
*GSTT1_MOUSE*
accession n°: Q64471
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 938.718 (0), 1587.95 (0), 1614.961 (0), 1624.93 (0),
1719.061 (0), 1742.999 (0), 1916.028 (0) }
Spot: 2D-0014AG (liver_mouse)
pI: 6.88 Mw: 25710
%vol: 0.0987723 %od: 0.0764068
================
*GSTT1_MOUSE*
accession n°: Q64471
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 938.718 (0), 1587.95 (0), 1614.961 (0), 1624.93 (0),
1719.061 (0), 1742.999 (0), 1916.028 (0) }
[Click to access protein entry]
Spot: 2D-0013M6 (liver_mouse)
pI: 6.58 Mw: 43547
%vol: 0.0822725 %od: 0.0846535
================
*FAAA_MOUSE*
accession n°: P35505
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 923.46 (0), 987.534 (0), 1070.574 (0), 1186.67 (0),
1289.624 (0), 1358.725 (0), 1653.952 (0), 1889.974 (0), 1938.925 (0),
1954.928 (0), 1970.911 (0), 2045.952 (0), 2086.088 (0), 2309.101 (0),
2366.098 (0), 2560.218 (0) }
Spot: 2D-0013M6 (liver_mouse)
pI: 6.58 Mw: 43547
%vol: 0.0822725 %od: 0.0846535
================
*FAAA_MOUSE*
accession n°: P35505
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 923.46 (0), 987.534 (0), 1070.574 (0), 1186.67 (0),
1289.624 (0), 1358.725 (0), 1653.952 (0), 1889.974 (0), 1938.925 (0),
1954.928 (0), 1970.911 (0), 2045.952 (0), 2086.088 (0), 2309.101 (0),
2366.098 (0), 2560.218 (0) }
[Click to access protein entry]
Spot: 2D-00148H (liver_mouse)
pI: 8.38 Mw: 27086
%vol: 0.120092 %od: 0.191266
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-00148H (liver_mouse)
pI: 8.38 Mw: 27086
%vol: 0.120092 %od: 0.191266
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0012PU (liver_mouse)
pI: 6.13 Mw: 121389
%vol: 0.0801178 %od: 0.0389608
================
*PYC_MOUSE*
accession n°: Q05920
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012PU (liver_mouse)
pI: 6.13 Mw: 121389
%vol: 0.0801178 %od: 0.0389608
================
*PYC_MOUSE*
accession n°: Q05920
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0013ET (liver_mouse)
pI: 7.09 Mw: 52035
%vol: 0.139994 %od: 0.308019
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013ET (liver_mouse)
pI: 7.09 Mw: 52035
%vol: 0.139994 %od: 0.308019
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0014LO (liver_mouse)
pI: 7.52 Mw: 12100
%vol: 0.161767 %od: 0.90177
================
*HBB1_MOUSE*
accession n°: P02088
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1126.565 (0), 1274.726 (0), 1294.648 (0), 1464.693 (0),
1730.013 (0), 1756.909 (0), 1996.893 (0) }
Spot: 2D-0014LO (liver_mouse)
pI: 7.52 Mw: 12100
%vol: 0.161767 %od: 0.90177
================
*HBB1_MOUSE*
accession n°: P02088
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1126.565 (0), 1274.726 (0), 1294.648 (0), 1464.693 (0),
1730.013 (0), 1756.909 (0), 1996.893 (0) }
[Click to access protein entry]
Spot: 2D-00146M (liver_mouse)
pI: 6.84 Mw: 27831
%vol: 0.157344 %od: 0.558887
================
*CAH3_MOUSE*
accession n°: P16015
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-00146M (liver_mouse)
pI: 6.84 Mw: 27831
%vol: 0.157344 %od: 0.558887
================
*CAH3_MOUSE*
accession n°: P16015
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013UV (liver_mouse)
pI: 6.80 Mw: 36427
%vol: 0.153091 %od: 0.244149
================
*OTC_MOUSE*
accession n°: P11725
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1117.637 (0), 1131.558 (0), 1161.595 (0), 1216.617 (0),
1278.663 (0), 1333.672 (0), 1429.718 (0), 1615.813 (0), 1672.831 (0),
1709.806 (0), 2108.941 (0), 2174.012 (0), 2445.234 (0), 2461.2 (0),
3698.849 (0) }
Spot: 2D-0013UV (liver_mouse)
pI: 6.80 Mw: 36427
%vol: 0.153091 %od: 0.244149
================
*OTC_MOUSE*
accession n°: P11725
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1117.637 (0), 1131.558 (0), 1161.595 (0), 1216.617 (0),
1278.663 (0), 1333.672 (0), 1429.718 (0), 1615.813 (0), 1672.831 (0),
1709.806 (0), 2108.941 (0), 2174.012 (0), 2445.234 (0), 2461.2 (0),
3698.849 (0) }
[Click to access protein entry]
Spot: 2D-0012NE (liver_mouse)
pI: 6.26 Mw: 158169
%vol: 0.129958 %od: 0.156531
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012NE (liver_mouse)
pI: 6.26 Mw: 158169
%vol: 0.129958 %od: 0.156531
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0014JV (liver_mouse)
pI: 4.42 Mw: 13856
%vol: 0.0350976 %od: 0.0591711
================
*RLA2_MOUSE*
accession n°: P99027
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014JV (liver_mouse)
pI: 4.42 Mw: 13856
%vol: 0.0350976 %od: 0.0591711
================
*RLA2_MOUSE*
accession n°: P99027
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0014ES (liver_mouse)
pI: 5.63 Mw: 21880
%vol: 0.0129277 %od: 0.0109733
================
*FRIL1_MOUSE*
accession n°: P29391
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 864.521 (0), 1034.535 (0), 1071.547 (0), 1461.744 (0),
1480.738 (0), 1686.859 (0), 1705.986 (0), 2373.269 (0)
}
Spot: 2D-0014ES (liver_mouse)
pI: 5.63 Mw: 21880
%vol: 0.0129277 %od: 0.0109733
================
*FRIL1_MOUSE*
accession n°: P29391
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 864.521 (0), 1034.535 (0), 1071.547 (0), 1461.744 (0),
1480.738 (0), 1686.859 (0), 1705.986 (0), 2373.269 (0)
}
[Click to access protein entry]
Spot: 2D-0013J9 (liver_mouse)
pI: 5.15 Mw: 46684
%vol: 0.0496129 %od: 0.0378817
================
*K1C18_MOUSE*
accession n°: P05784
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013J9 (liver_mouse)
pI: 5.15 Mw: 46684
%vol: 0.0496129 %od: 0.0378817
================
*K1C18_MOUSE*
accession n°: P05784
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013BS (liver_mouse)
pI: 6.85 Mw: 55629
%vol: 0.121056 %od: 0.290778
================
*CATA_MOUSE*
accession n°: P24270
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 829.452 (0), 888.431 (0), 912.457 (0), 1119.559 (0),
1276.606 (0), 1294.568 (0), 1391.633 (0), 1493.692 (0), 1502.807 (0),
1655.798 (0), 1740.831 (0), 1803.929 (0), 1882.969 (0), 2205.065 (0)
}
Spot: 2D-0013BS (liver_mouse)
pI: 6.85 Mw: 55629
%vol: 0.121056 %od: 0.290778
================
*CATA_MOUSE*
accession n°: P24270
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 829.452 (0), 888.431 (0), 912.457 (0), 1119.559 (0),
1276.606 (0), 1294.568 (0), 1391.633 (0), 1493.692 (0), 1502.807 (0),
1655.798 (0), 1740.831 (0), 1803.929 (0), 1882.969 (0), 2205.065 (0)
}
[Click to access protein entry]
Spot: 2D-001480 (liver_mouse)
pI: 5.33 Mw: 27370
%vol: 0.0129844 %od: 0.00507947
================
*GAMT_MOUSE*
accession n°: O35969
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 901.485 (0), 1045.554 (0), 1420.751 (0), 1436.794 (0),
1687.923 (0), 1761.901 (0), 2045.938 (0), 2568.323 (0), 2648.419 (0)
}
Spot: 2D-001480 (liver_mouse)
pI: 5.33 Mw: 27370
%vol: 0.0129844 %od: 0.00507947
================
*GAMT_MOUSE*
accession n°: O35969
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 901.485 (0), 1045.554 (0), 1420.751 (0), 1436.794 (0),
1687.923 (0), 1761.901 (0), 2045.938 (0), 2568.323 (0), 2648.419 (0)
}
[Click to access protein entry]
Spot: 2D-0013Y3 (liver_mouse)
pI: 6.98 Mw: 34000
%vol: 0.12236 %od: 0.257228
================
*LDHA_MOUSE*
accession n°: P06151
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 913.586 (0), 1118.557 (0), 1144.598 (0), 1197.632 (0),
1248.635 (0), 1830.917 (0), 1931.991 (0), 1944.086 (0), 1964.979 (0)
}
Spot: 2D-0013Y3 (liver_mouse)
pI: 6.98 Mw: 34000
%vol: 0.12236 %od: 0.257228
================
*LDHA_MOUSE*
accession n°: P06151
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 913.586 (0), 1118.557 (0), 1144.598 (0), 1197.632 (0),
1248.635 (0), 1830.917 (0), 1931.991 (0), 1944.086 (0), 1964.979 (0)
}
[Click to access protein entry]
Spot: 2D-0014IC (liver_mouse)
pI: 7.18 Mw: 15283
%vol: 0.100984 %od: 0.167722
================
*PPIA_MOUSE*
accession n°: P17742
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1055.464 (0), 1154.504 (0), 1310.527 (0), 1379.728 (0),
1614.688 (0), 1831.821 (0) }
Spot: 2D-0014IC (liver_mouse)
pI: 7.18 Mw: 15283
%vol: 0.100984 %od: 0.167722
================
*PPIA_MOUSE*
accession n°: P17742
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1055.464 (0), 1154.504 (0), 1310.527 (0), 1379.728 (0),
1614.688 (0), 1831.821 (0) }
[Click to access protein entry]
Spot: 2D-0013B7 (liver_mouse)
pI: 7.26 Mw: 56566
%vol: 0.119638 %od: 0.292349
================
*CATA_MOUSE*
accession n°: P24270
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 888.421 (0), 912.481 (0), 962.45 (0), 1119.526 (0),
1154.521 (0), 1276.602 (0), 1294.536 (0), 1391.605 (0), 1493.677 (0),
1502.776 (0), 1542.785 (0), 1655.778 (0), 1716.753 (0), 1740.805 (0),
1803.913 (0), 1882.928 (0), 2205.071 (0) }
Spot: 2D-0013B7 (liver_mouse)
pI: 7.26 Mw: 56566
%vol: 0.119638 %od: 0.292349
================
*CATA_MOUSE*
accession n°: P24270
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 888.421 (0), 912.481 (0), 962.45 (0), 1119.526 (0),
1154.521 (0), 1276.602 (0), 1294.536 (0), 1391.605 (0), 1493.677 (0),
1502.776 (0), 1542.785 (0), 1655.778 (0), 1716.753 (0), 1740.805 (0),
1803.913 (0), 1882.928 (0), 2205.071 (0) }
[Click to access protein entry]
Spot: 2D-0014IR (liver_mouse)
pI: 4.88 Mw: 14567
%vol: 0.10671 %od: 0.235272
================
*CYB5_MOUSE*
accession n°: P56395
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1120.64 (0), 1186.61 (0), 1428.703 (0), 1511.755 (0),
2205.941 (0) }
Spot: 2D-0014IR (liver_mouse)
pI: 4.88 Mw: 14567
%vol: 0.10671 %od: 0.235272
================
*CYB5_MOUSE*
accession n°: P56395
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1120.64 (0), 1186.61 (0), 1428.703 (0), 1511.755 (0),
2205.941 (0) }
[Click to access protein entry]
Spot: 2D-0013EJ (liver_mouse)
pI: 7.30 Mw: 52035
%vol: 0.15604 %od: 0.496097
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013EJ (liver_mouse)
pI: 7.30 Mw: 52035
%vol: 0.15604 %od: 0.496097
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0014LZ (liver_mouse)
pI: 6.33 Mw: 11946
%vol: 0.119808 %od: 0.25438
================
*DOPD_MOUSE*
accession n°: O35215
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 869.468 (0), 924.448 (0), 1335.708 (0), 1465.729 (0),
1472.785 (0), 1673.868 (0), 1691.913 (0), 1947.029 (0), 2425.211 (0)
}
Spot: 2D-0014LZ (liver_mouse)
pI: 6.33 Mw: 11946
%vol: 0.119808 %od: 0.25438
================
*DOPD_MOUSE*
accession n°: O35215
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 869.468 (0), 924.448 (0), 1335.708 (0), 1465.729 (0),
1472.785 (0), 1673.868 (0), 1691.913 (0), 1947.029 (0), 2425.211 (0)
}
[Click to access protein entry]
Spot: 2D-001491 (liver_mouse)
pI: 6.94 Mw: 26694
%vol: 0.147308 %od: 0.254049
================
*TPIS_MOUSE*
accession n°: P17751
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 954.474 (0), 1082.583 (0), 1247.607 (0), 1458.722 (0),
1466.705 (0), 1539.799 (0), 1602.868 (0), 1614.844 (0), 1621.859 (0),
1837.96 (0) }
Spot: 2D-001491 (liver_mouse)
pI: 6.94 Mw: 26694
%vol: 0.147308 %od: 0.254049
================
*TPIS_MOUSE*
accession n°: P17751
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 954.474 (0), 1082.583 (0), 1247.607 (0), 1458.722 (0),
1466.705 (0), 1539.799 (0), 1602.868 (0), 1614.844 (0), 1621.859 (0),
1837.96 (0) }
[Click to access protein entry]
Spot: 2D-0014B9 (liver_mouse)
pI: 7.83 Mw: 24970
%vol: 0.17821 %od: 0.883049
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0014B9 (liver_mouse)
pI: 7.83 Mw: 24970
%vol: 0.17821 %od: 0.883049
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013BF (liver_mouse)
pI: 5.84 Mw: 56408
%vol: 0.143566 %od: 0.294153
================
*PDIA3_MOUSE*
accession n°: P27773
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 877.494 (0), 995.562 (0), 997.514 (0), 1084.565 (0),
1172.552 (0), 1188.532 (0), 1191.589 (0), 1258.668 (0), 1341.693 (0),
1382.678 (0), 1394.652 (0), 1593.83 (0), 1607.749 (0), 1653.743 (0),
1814.891 (0), 2302.11 (0) }
Spot: 2D-0013BF (liver_mouse)
pI: 5.84 Mw: 56408
%vol: 0.143566 %od: 0.294153
================
*PDIA3_MOUSE*
accession n°: P27773
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 877.494 (0), 995.562 (0), 997.514 (0), 1084.565 (0),
1172.552 (0), 1188.532 (0), 1191.589 (0), 1258.668 (0), 1341.693 (0),
1382.678 (0), 1394.652 (0), 1593.83 (0), 1607.749 (0), 1653.743 (0),
1814.891 (0), 2302.11 (0) }
[Click to access protein entry]
Spot: 2D-0013LB (liver_mouse)
pI: 5.95 Mw: 44527
%vol: 0.0728035 %od: 0.0425975
================
*ODBA_MOUSE*
accession n°: P50136
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 831.403 (0), 908.501 (0), 954.442 (0), 1098.551 (0),
1225.56 (0), 1512.717 (0), 1532.667 (0), 1700.777 (0), 1868.914 (0),
2441.327 (0), 2519.136 (0) }
Spot: 2D-0013LB (liver_mouse)
pI: 5.95 Mw: 44527
%vol: 0.0728035 %od: 0.0425975
================
*ODBA_MOUSE*
accession n°: P50136
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 831.403 (0), 908.501 (0), 954.442 (0), 1098.551 (0),
1225.56 (0), 1512.717 (0), 1532.667 (0), 1700.777 (0), 1868.914 (0),
2441.327 (0), 2519.136 (0) }
[Click to access protein entry]
Spot: 2D-0014C6 (liver_mouse)
pI: 5.46 Mw: 24660
%vol: 0.0242112 %od: 0.0199491
================
*PSB4_MOUSE*
accession n°: P99026
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014C6 (liver_mouse)
pI: 5.46 Mw: 24660
%vol: 0.0242112 %od: 0.0199491
================
*PSB4_MOUSE*
accession n°: P99026
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-00148W (liver_mouse)
pI: 4.72 Mw: 26694
%vol: 0.0697417 %od: 0.071203
================
*COX2_MOUSE*
accession n°: P00405
Identification Methods:
* MAPPING: {Gm}
================
*TPM3_MOUSE*
accession n: P21107
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-00148W (liver_mouse)
pI: 4.72 Mw: 26694
%vol: 0.0697417 %od: 0.071203
================
*COX2_MOUSE*
accession n°: P00405
Identification Methods:
* MAPPING: {Gm}
================
*TPM3_MOUSE*
accession n: P21107
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-001343 (liver_mouse)
pI: 5.69 Mw: 66707
%vol: 0.0342471 %od: 0.0216479
================
*ALBU_MOUSE*
accession n°: P07724
Identification Methods:
* MAPPING: {Mi,Gm}
Spot: 2D-001343 (liver_mouse)
pI: 5.69 Mw: 66707
%vol: 0.0342471 %od: 0.0216479
================
*ALBU_MOUSE*
accession n°: P07724
Identification Methods:
* MAPPING: {Mi,Gm}
[Click to access protein entry]
Spot: 2D-0013XR (liver_mouse)
pI: 8.09 Mw: 34228
%vol: 0.141808 %od: 0.181135
================
*URIC_MOUSE*
accession n°: P25688
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1007.491 (0), 1118.635 (0), 1177.63 (0), 1214.619 (0),
1216.665 (0), 1356.706 (0), 1377.681 (0), 1486.763 (0), 1916.951 (0)
}
Spot: 2D-0013XR (liver_mouse)
pI: 8.09 Mw: 34228
%vol: 0.141808 %od: 0.181135
================
*URIC_MOUSE*
accession n°: P25688
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1007.491 (0), 1118.635 (0), 1177.63 (0), 1214.619 (0),
1216.665 (0), 1356.706 (0), 1377.681 (0), 1486.763 (0), 1916.951 (0)
}
[Click to access protein entry]
Spot: 2D-0012UL (liver_mouse)
pI: 5.43 Mw: 90604
%vol: 0.0175772 %od: 0.0107958
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012UL (liver_mouse)
pI: 5.43 Mw: 90604
%vol: 0.0175772 %od: 0.0107958
================
*FTDH_MOUSE*
accession n°: P99017
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-00133U (liver_mouse)
pI: 5.30 Mw: 67063
%vol: 0.130128 %od: 0.366604
================
*HSP7C_MOUSE*
accession n°: P63017
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1197.663 (0), 1199.7 (0), 1228.612 (0), 1253.626 (0),
1487.703 (0), 1981.974 (0) }
Spot: 2D-00133U (liver_mouse)
pI: 5.30 Mw: 67063
%vol: 0.130128 %od: 0.366604
================
*HSP7C_MOUSE*
accession n°: P63017
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1197.663 (0), 1199.7 (0), 1228.612 (0), 1253.626 (0),
1487.703 (0), 1981.974 (0) }
[Click to access protein entry]
Spot: 2D-001331 (liver_mouse)
pI: 5.32 Mw: 69057
%vol: 0.085958 %od: 0.0670319
================
*GRP75_MOUSE*
accession n°: P38647
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1242.688 (0), 1290.676 (0), 1450.713 (0), 1462.791 (0),
1569.772 (0), 1592.949 (0), 1645.937 (0), 1694.862 (0), 1808.887 (0),
2055.953 (0), 2251.182 (0), 2309.26 (0) }
Spot: 2D-001331 (liver_mouse)
pI: 5.32 Mw: 69057
%vol: 0.085958 %od: 0.0670319
================
*GRP75_MOUSE*
accession n°: P38647
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1242.688 (0), 1290.676 (0), 1450.713 (0), 1462.791 (0),
1569.772 (0), 1592.949 (0), 1645.937 (0), 1694.862 (0), 1808.887 (0),
2055.953 (0), 2251.182 (0), 2309.26 (0) }
[Click to access protein entry]
Spot: 2D-0013F6 (liver_mouse)
pI: 6.77 Mw: 51174
%vol: 0.146854 %od: 0.451258
================
*DHE3_MOUSE*
accession n°: P26443
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013F6 (liver_mouse)
pI: 6.77 Mw: 51174
%vol: 0.146854 %od: 0.451258
================
*DHE3_MOUSE*
accession n°: P26443
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013U9 (liver_mouse)
pI: 6.85 Mw: 37164
%vol: 0.148725 %od: 0.155554
================
*ALDX_MOUSE*
accession n°: P99052
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 897.572 (0), 1505.67 (0), 1551.742 (0), 2098.048 (0),
2201.997 (0) }
Spot: 2D-0013U9 (liver_mouse)
pI: 6.85 Mw: 37164
%vol: 0.148725 %od: 0.155554
================
*ALDX_MOUSE*
accession n°: P99052
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 897.572 (0), 1505.67 (0), 1551.742 (0), 2098.048 (0),
2201.997 (0) }
[Click to access protein entry]
Spot: 2D-00143C (liver_mouse)
pI: 6.59 Mw: 30315
%vol: 0.0554532 %od: 0.0610065
================
*GSTA3_MOUSE*
accession n°: P30115
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 901.45 (0), 932.526 (0), 1060.612 (0), 1245.626 (0),
1401.758 (0), 1462.793 (0), 1494.709 (0), 2160.021 (0)
}
Spot: 2D-00143C (liver_mouse)
pI: 6.59 Mw: 30315
%vol: 0.0554532 %od: 0.0610065
================
*GSTA3_MOUSE*
accession n°: P30115
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 901.45 (0), 932.526 (0), 1060.612 (0), 1245.626 (0),
1401.758 (0), 1462.793 (0), 1494.709 (0), 2160.021 (0)
}
[Click to access protein entry]
Spot: 2D-00148U (liver_mouse)
pI: 7.37 Mw: 26861
%vol: 0.0675303 %od: 0.110843
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1015.579 (0), 1105.58 (0), 1248.672 (0), 1264.649 (0),
1899.009 (0), 1993.997 (0) }
Spot: 2D-00148U (liver_mouse)
pI: 7.37 Mw: 26861
%vol: 0.0675303 %od: 0.110843
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1015.579 (0), 1105.58 (0), 1248.672 (0), 1264.649 (0),
1899.009 (0), 1993.997 (0) }
[Click to access protein entry]
Spot: 2D-0013B9 (liver_mouse)
pI: 4.43 Mw: 56566
%vol: 0.0345873 %od: 0.0229046
================
*CALR_MOUSE*
accession n°: P14211
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 868.445 (0), 871.455 (0), 975.491 (0), 992.472 (0),
1004.544 (0), 1019.581 (0), 1219.719 (0), 1451.677 (0)
}
Spot: 2D-0013B9 (liver_mouse)
pI: 4.43 Mw: 56566
%vol: 0.0345873 %od: 0.0229046
================
*CALR_MOUSE*
accession n°: P14211
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 868.445 (0), 871.455 (0), 975.491 (0), 992.472 (0),
1004.544 (0), 1019.581 (0), 1219.719 (0), 1451.677 (0)
}
[Click to access protein entry]
Spot: 2D-00133T (liver_mouse)
pI: 5.28 Mw: 67421
%vol: 0.0967878 %od: 0.0659648
================
*HSP7C_MOUSE*
accession n°: P63017
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 993.505 (0), 1081.531 (0), 1197.649 (0), 1199.693 (0),
1228.655 (0), 1253.63 (0), 1481.815 (0), 1487.722 (0), 1659.93 (0),
1691.75 (0), 1982.021 (0) }
Spot: 2D-00133T (liver_mouse)
pI: 5.28 Mw: 67421
%vol: 0.0967878 %od: 0.0659648
================
*HSP7C_MOUSE*
accession n°: P63017
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 993.505 (0), 1081.531 (0), 1197.649 (0), 1199.693 (0),
1228.655 (0), 1253.63 (0), 1481.815 (0), 1487.722 (0), 1659.93 (0),
1691.75 (0), 1982.021 (0) }
[Click to access protein entry]
Spot: 2D-0014BG (liver_mouse)
pI: 7.09 Mw: 24866
%vol: 0.152468 %od: 0.384165
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0014BG (liver_mouse)
pI: 7.09 Mw: 24866
%vol: 0.152468 %od: 0.384165
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013IW (liver_mouse)
pI: 6.34 Mw: 46684
%vol: 0.153942 %od: 0.46747
================
*ENOA_MOUSE*
accession n°: P17182
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1143.5 (0), 1181.5 (0), 1439.7 (0), 1804.9 (0), 1928.8 (0)
}
Spot: 2D-0013IW (liver_mouse)
pI: 6.34 Mw: 46684
%vol: 0.153942 %od: 0.46747
================
*ENOA_MOUSE*
accession n°: P17182
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1143.5 (0), 1181.5 (0), 1439.7 (0), 1804.9 (0), 1928.8 (0)
}
[Click to access protein entry]
Spot: 2D-0013W8 (liver_mouse)
pI: 8.35 Mw: 35626
%vol: 0.137102 %od: 0.216134
================
*MDHM_MOUSE*
accession n°: P08249
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 992.591 (0), 1147.71 (0), 1233.715 (0), 1338.734 (0),
1489.769 (0), 1560.81 (0), 1793.12 (0), 2393.241 (0)
}
Spot: 2D-0013W8 (liver_mouse)
pI: 8.35 Mw: 35626
%vol: 0.137102 %od: 0.216134
================
*MDHM_MOUSE*
accession n°: P08249
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 992.591 (0), 1147.71 (0), 1233.715 (0), 1338.734 (0),
1489.769 (0), 1560.81 (0), 1793.12 (0), 2393.241 (0)
}
[Click to access protein entry]
Spot: 2D-00138B (liver_mouse)
pI: 4.44 Mw: 59607
%vol: 0.110509 %od: 0.172741
================
*CALR_MOUSE*
accession n°: P14211
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 853.461 (0), 868.423 (0), 871.428 (0), 975.501 (0), 978.406 (0),
992.478 (0), 1004.543 (0), 1019.566 (0), 1219.709 (0), 1451.669 (0)
}
Spot: 2D-00138B (liver_mouse)
pI: 4.44 Mw: 59607
%vol: 0.110509 %od: 0.172741
================
*CALR_MOUSE*
accession n°: P14211
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 853.461 (0), 868.423 (0), 871.428 (0), 975.501 (0), 978.406 (0),
992.478 (0), 1004.543 (0), 1019.566 (0), 1219.709 (0), 1451.669 (0)
}
[Click to access protein entry]
Spot: 2D-00131W (liver_mouse)
pI: 5.26 Mw: 71299
%vol: 0.0225101 %od: 0.00757054
================
*NCPR_MOUSE*
accession n°: P37040
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-00131W (liver_mouse)
pI: 5.26 Mw: 71299
%vol: 0.0225101 %od: 0.00757054
================
*NCPR_MOUSE*
accession n°: P37040
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013MQ (liver_mouse)
pI: 8.03 Mw: 43185
%vol: 0.140731 %od: 0.132235
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013MQ (liver_mouse)
pI: 8.03 Mw: 43185
%vol: 0.140731 %od: 0.132235
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013WE (liver_mouse)
pI: 8.14 Mw: 35389
%vol: 0.158988 %od: 0.2414
================
*DHB5_MOUSE*
accession n°: P70694
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 854.545 (0), 862.468 (0), 993.525 (0), 1158.578 (0),
1467.619 (0), 1500.828 (0), 1599.814 (0), 1715.954 (0), 2234.046 (0),
2386.235 (0) }
Spot: 2D-0013WE (liver_mouse)
pI: 8.14 Mw: 35389
%vol: 0.158988 %od: 0.2414
================
*DHB5_MOUSE*
accession n°: P70694
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 854.545 (0), 862.468 (0), 993.525 (0), 1158.578 (0),
1467.619 (0), 1500.828 (0), 1599.814 (0), 1715.954 (0), 2234.046 (0),
2386.235 (0) }
[Click to access protein entry]
Spot: 2D-0013F2 (liver_mouse)
pI: 6.56 Mw: 51747
%vol: 0.0525046 %od: 0.0726815
================
*DHE3_MOUSE*
accession n°: P26443
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.535 (0), 1059.549 (0), 1425.624 (0), 1723.869 (0),
1764.871 (0), 1931.923 (0), 1936.893 (0) }
Spot: 2D-0013F2 (liver_mouse)
pI: 6.56 Mw: 51747
%vol: 0.0525046 %od: 0.0726815
================
*DHE3_MOUSE*
accession n°: P26443
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.535 (0), 1059.549 (0), 1425.624 (0), 1723.869 (0),
1764.871 (0), 1931.923 (0), 1936.893 (0) }
[Click to access protein entry]
Spot: 2D-0012PV (liver_mouse)
pI: 6.21 Mw: 120000
%vol: 0.12786 %od: 0.200872
================
*PYC_MOUSE*
accession n°: Q05920
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1146.681 (0), 1321.837 (0), 1350.704 (0), 1654.785 (0),
1767.76 (0), 1993.98 (0) }
Spot: 2D-0012PV (liver_mouse)
pI: 6.21 Mw: 120000
%vol: 0.12786 %od: 0.200872
================
*PYC_MOUSE*
accession n°: Q05920
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1146.681 (0), 1321.837 (0), 1350.704 (0), 1654.785 (0),
1767.76 (0), 1993.98 (0) }
[Click to access protein entry]
Spot: 2D-0013PN (liver_mouse)
pI: 5.28 Mw: 40532
%vol: 0.0174638 %od: 0.0161023
================
*KTRA_MOUSE*
accession n°: P99018
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013PN (liver_mouse)
pI: 5.28 Mw: 40532
%vol: 0.0174638 %od: 0.0161023
================
*KTRA_MOUSE*
accession n°: P99018
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0012UJ (liver_mouse)
pI: 4.89 Mw: 90000
%vol: 0.0909477 %od: 0.0251139
================
*ENPL_MOUSE*
accession n°: P08113
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0012UJ (liver_mouse)
pI: 4.89 Mw: 90000
%vol: 0.0909477 %od: 0.0251139
================
*ENPL_MOUSE*
accession n°: P08113
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-00136H (liver_mouse)
pI: 6.84 Mw: 63113
%vol: 0.0366285 %od: 0.0238607
================
*MAOX_MOUSE*
accession n°: P06801
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 866.349 (0), 959.476 (0), 1086.646 (0), 1093.542 (0),
1295.673 (0), 1846.911 (0), 2211.104 (0) }
Spot: 2D-00136H (liver_mouse)
pI: 6.84 Mw: 63113
%vol: 0.0366285 %od: 0.0238607
================
*MAOX_MOUSE*
accession n°: P06801
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 866.349 (0), 959.476 (0), 1086.646 (0), 1093.542 (0),
1295.673 (0), 1846.911 (0), 2211.104 (0) }
[Click to access protein entry]
Spot: 2D-0013NC (liver_mouse)
pI: 7.32 Mw: 42707
%vol: 0.107334 %od: 0.0413375
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013NC (liver_mouse)
pI: 7.32 Mw: 42707
%vol: 0.107334 %od: 0.0413375
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-00133R (liver_mouse)
pI: 5.25 Mw: 67781
%vol: 0.031639 %od: 0.0272874
================
*HSP7C_MOUSE*
accession n°: P63017
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-00133R (liver_mouse)
pI: 5.25 Mw: 67781
%vol: 0.031639 %od: 0.0272874
================
*HSP7C_MOUSE*
accession n°: P63017
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013EA (liver_mouse)
pI: 5.63 Mw: 52764
%vol: 0.126442 %od: 0.217307
================
*SBP1_MOUSE*
accession n°: P17563
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013EA (liver_mouse)
pI: 5.63 Mw: 52764
%vol: 0.126442 %od: 0.217307
================
*SBP1_MOUSE*
accession n°: P17563
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013MP (liver_mouse)
pI: 6.75 Mw: 42945
%vol: 0.162164 %od: 0.317065
================
*GLNA_MOUSE*
accession n°: P15105
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1146.562 (0), 1810.877 (0), 1918.943 (0), 2150.054 (0),
2720.325 (0) }
Spot: 2D-0013MP (liver_mouse)
pI: 6.75 Mw: 42945
%vol: 0.162164 %od: 0.317065
================
*GLNA_MOUSE*
accession n°: P15105
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1146.562 (0), 1810.877 (0), 1918.943 (0), 2150.054 (0),
2720.325 (0) }
[Click to access protein entry]
Spot: 2D-00149S (liver_mouse)
pI: 5.90 Mw: 26197
%vol: 0.0233039 %od: 0.0250712
================
*PRDX6_MOUSE*
accession n°: O08709
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 906.463 (0), 1021.556 (0), 1045.548 (0), 1057.536 (0),
1149.637 (0), 1191.641 (0), 1395.607 (0), 1896.859 (0), 2030.865 (0),
2141.948 (0), 2150.954 (0) }
Spot: 2D-00149S (liver_mouse)
pI: 5.90 Mw: 26197
%vol: 0.0233039 %od: 0.0250712
================
*PRDX6_MOUSE*
accession n°: O08709
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 906.463 (0), 1021.556 (0), 1045.548 (0), 1057.536 (0),
1149.637 (0), 1191.641 (0), 1395.607 (0), 1896.859 (0), 2030.865 (0),
2141.948 (0), 2150.954 (0) }
[Click to access protein entry]
Spot: 2D-00134R (liver_mouse)
pI: 5.50 Mw: 65673
%vol: 0.137726 %od: 0.205916
================
*ALBU_MOUSE*
accession n°: P07724
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 956.51 (0), 1149.594 (0), 1250.625 (0), 1299.714 (0),
1439.795 (0), 1479.786 (0), 1609.775 (0), 1662.832 (0), 1681.867 (0),
1809.714 (0), 1825.96 (0), 1838.939 (0), 1882.918 (0), 1896.005 (0),
1917.997 (0) }
Spot: 2D-00134R (liver_mouse)
pI: 5.50 Mw: 65673
%vol: 0.137726 %od: 0.205916
================
*ALBU_MOUSE*
accession n°: P07724
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 956.51 (0), 1149.594 (0), 1250.625 (0), 1299.714 (0),
1439.795 (0), 1479.786 (0), 1609.775 (0), 1662.832 (0), 1681.867 (0),
1809.714 (0), 1825.96 (0), 1838.939 (0), 1882.918 (0), 1896.005 (0),
1917.997 (0) }
[Click to access protein entry]
Spot: 2D-0014IH (liver_mouse)
pI: 7.60 Mw: 15061
%vol: 0.0821024 %od: 0.133205
================
*PPIA_MOUSE*
accession n°: P17742
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1055.53 (0), 1154.597 (0), 1614.73 (0), 2005.949 (0)
}
Spot: 2D-0014IH (liver_mouse)
pI: 7.60 Mw: 15061
%vol: 0.0821024 %od: 0.133205
================
*PPIA_MOUSE*
accession n°: P17742
Identification Methods:
* MAPPING: {Mi} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1055.53 (0), 1154.597 (0), 1614.73 (0), 2005.949 (0)
}
[Click to access protein entry]
Spot: 2D-00139R (liver_mouse)
pI: 5.26 Mw: 58289
%vol: 0.0612933 %od: 0.0497805
================
*CH60_MOUSE*
accession n°: P63038
Identification Methods:
* MAPPING: {Mi,Gm}
Spot: 2D-00139R (liver_mouse)
pI: 5.26 Mw: 58289
%vol: 0.0612933 %od: 0.0497805
================
*CH60_MOUSE*
accession n°: P63038
Identification Methods:
* MAPPING: {Mi,Gm}
[Click to access protein entry]
Spot: 2D-0013C7 (liver_mouse)
pI: 6.78 Mw: 55320
%vol: 0.102344 %od: 0.143697
================
*CATA_MOUSE*
accession n°: P24270
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1119.55 (0), 1276.616 (0), 1294.522 (0), 1391.625 (0),
1493.686 (0), 1502.817 (0), 1655.778 (0), 1740.826 (0), 1803.901 (0),
2205.084 (0) }
Spot: 2D-0013C7 (liver_mouse)
pI: 6.78 Mw: 55320
%vol: 0.102344 %od: 0.143697
================
*CATA_MOUSE*
accession n°: P24270
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1119.55 (0), 1276.616 (0), 1294.522 (0), 1391.625 (0),
1493.686 (0), 1502.817 (0), 1655.778 (0), 1740.826 (0), 1803.901 (0),
2205.084 (0) }
[Click to access protein entry]
Spot: 2D-0012T7 (liver_mouse)
pI: 6.68 Mw: 99501
%vol: 0.110679 %od: 0.101287
================
*ACOC_MOUSE*
accession n°: P28271
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 889.545 (0), 1050.537 (0), 1100.591 (0), 1170.655 (0),
1458.784 (0), 1599.842 (0), 1852.897 (0), 2060.067 (0), 2090.097 (0)
}
Spot: 2D-0012T7 (liver_mouse)
pI: 6.68 Mw: 99501
%vol: 0.110679 %od: 0.101287
================
*ACOC_MOUSE*
accession n°: P28271
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 889.545 (0), 1050.537 (0), 1100.591 (0), 1170.655 (0),
1458.784 (0), 1599.842 (0), 1852.897 (0), 2060.067 (0), 2090.097 (0)
}
[Click to access protein entry]
Spot: 2D-0013NP (liver_mouse)
pI: 5.27 Mw: 42117
%vol: 0.109149 %od: 0.0642403
================
*ACTB_MOUSE*
accession n°: P99041
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013NP (liver_mouse)
pI: 5.27 Mw: 42117
%vol: 0.109149 %od: 0.0642403
================
*ACTB_MOUSE*
accession n°: P99041
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-00132L (liver_mouse)
pI: 5.01 Mw: 70169
%vol: 0.107051 %od: 0.0733815
================
*GRP78_MOUSE*
accession n°: P20029
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-00132L (liver_mouse)
pI: 5.01 Mw: 70169
%vol: 0.107051 %od: 0.0733815
================
*GRP78_MOUSE*
accession n°: P20029
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013XP (liver_mouse)
pI: 8.85 Mw: 34304
%vol: 0.0567572 %od: 0.0469753
================
*ROA2_MOUSE*
accession n°: O88569
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1013.571 (0), 1377.693 (0), 1410.742 (0), 1798.924 (0),
2205.901 (0), 2495.035 (0) }
Spot: 2D-0013XP (liver_mouse)
pI: 8.85 Mw: 34304
%vol: 0.0567572 %od: 0.0469753
================
*ROA2_MOUSE*
accession n°: O88569
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1013.571 (0), 1377.693 (0), 1410.742 (0), 1798.924 (0),
2205.901 (0), 2495.035 (0) }
[Click to access protein entry]
Spot: 2D-0013E4 (liver_mouse)
pI: 5.13 Mw: 53059
%vol: 0.0572675 %od: 0.0255852
================
*VIME_MOUSE*
accession n°: P20152
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013E4 (liver_mouse)
pI: 5.13 Mw: 53059
%vol: 0.0572675 %od: 0.0255852
================
*VIME_MOUSE*
accession n°: P20152
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0014M4 (liver_mouse)
pI: 6.86 Mw: 11838
%vol: 0.068721 %od: 0.115791
================
*CX6A1_MOUSE*
accession n°: P43024
Identification Methods:
* MAPPING: {Mi,PMF}
peptide masses: { (TRYPSIN)
$m/Z= 2014.016 (0), 2255.261 (0), 2283.204 (0), 3316.568 (0)
}
Spot: 2D-0014M4 (liver_mouse)
pI: 6.86 Mw: 11838
%vol: 0.068721 %od: 0.115791
================
*CX6A1_MOUSE*
accession n°: P43024
Identification Methods:
* MAPPING: {Mi,PMF}
peptide masses: { (TRYPSIN)
$m/Z= 2014.016 (0), 2255.261 (0), 2283.204 (0), 3316.568 (0)
}
[Click to access protein entry]
Spot: 2D-001330 (liver_mouse)
pI: 5.27 Mw: 69426
%vol: 0.0199586 %od: 0.0108828
================
*GRP75_MOUSE*
accession n°: P38647
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
Spot: 2D-001330 (liver_mouse)
pI: 5.27 Mw: 69426
%vol: 0.0199586 %od: 0.0108828
================
*GRP75_MOUSE*
accession n°: P38647
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
[Click to access protein entry]
Spot: 2D-0013M5 (liver_mouse)
pI: 6.31 Mw: 43426
%vol: 0.0661128 %od: 0.0462308
================
*ODBA_MOUSE*
accession n°: P50136
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013M5 (liver_mouse)
pI: 6.31 Mw: 43426
%vol: 0.0661128 %od: 0.0462308
================
*ODBA_MOUSE*
accession n°: P50136
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013GM (liver_mouse)
pI: 5.05 Mw: 49082
%vol: 0.146968 %od: 0.447384
================
*ATPB_MOUSE*
accession n°: P56480
Identification Methods:
* MAPPING: {Mi,Gm}
Spot: 2D-0013GM (liver_mouse)
pI: 5.05 Mw: 49082
%vol: 0.146968 %od: 0.447384
================
*ATPB_MOUSE*
accession n°: P56480
Identification Methods:
* MAPPING: {Mi,Gm}
[Click to access protein entry]
Spot: 2D-0014LD (liver_mouse)
pI: 6.02 Mw: 12511
%vol: 0.0222833 %od: 0.0239068
================
*ULAK_MOUSE*
accession n°: P99031
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014LD (liver_mouse)
pI: 6.02 Mw: 12511
%vol: 0.0222833 %od: 0.0239068
================
*ULAK_MOUSE*
accession n°: P99031
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0012OH (liver_mouse)
pI: 5.85 Mw: 140977
%vol: 0.0406544 %od: 0.0148765
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
Spot: 2D-0012OH (liver_mouse)
pI: 5.85 Mw: 140977
%vol: 0.0406544 %od: 0.0148765
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
[Click to access protein entry]
Spot: 2D-0013E1 (liver_mouse)
pI: 4.43 Mw: 52618
%vol: 0.0179174 %od: 0.0153408
================
*CALR_MOUSE*
accession n°: P14211
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0013E1 (liver_mouse)
pI: 4.43 Mw: 52618
%vol: 0.0179174 %od: 0.0153408
================
*CALR_MOUSE*
accession n°: P14211
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0014LC (liver_mouse)
pI: 9.08 Mw: 12485
%vol: 0.104216 %od: 0.174641
================
*UK114_MOUSE*
accession n°: P52760
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1059.608 (0), 1137.573 (0), 1139.591 (0), 1513.858 (0),
1642.865 (0), 2144.067 (0), 2160.025 (0), 2272.07 (0), 2700.333 (0)
}
Spot: 2D-0014LC (liver_mouse)
pI: 9.08 Mw: 12485
%vol: 0.104216 %od: 0.174641
================
*UK114_MOUSE*
accession n°: P52760
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1059.608 (0), 1137.573 (0), 1139.591 (0), 1513.858 (0),
1642.865 (0), 2144.067 (0), 2160.025 (0), 2272.07 (0), 2700.333 (0)
}
[Click to access protein entry]
Spot: 2D-0013M7 (liver_mouse)
pI: 8.26 Mw: 43426
%vol: 0.107221 %od: 0.120473
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013M7 (liver_mouse)
pI: 8.26 Mw: 43426
%vol: 0.107221 %od: 0.120473
================
*THIM_MOUSE*
accession n°: P99023
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013DY (liver_mouse)
pI: 5.70 Mw: 53059
%vol: 0.107731 %od: 0.122969
================
*K2C8_MOUSE*
accession n°: P11679
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013DY (liver_mouse)
pI: 5.70 Mw: 53059
%vol: 0.107731 %od: 0.122969
================
*K2C8_MOUSE*
accession n°: P11679
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0014BB (liver_mouse)
pI: 8.07 Mw: 25127
%vol: 0.157117 %od: 0.239467
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0014BB (liver_mouse)
pI: 8.07 Mw: 25127
%vol: 0.157117 %od: 0.239467
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0014LU (liver_mouse)
pI: 4.99 Mw: 12075
%vol: 0.0328296 %od: 0.0417763
================
*QCR6_MOUSE*
accession n°: P99028
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014LU (liver_mouse)
pI: 4.99 Mw: 12075
%vol: 0.0328296 %od: 0.0417763
================
*QCR6_MOUSE*
accession n°: P99028
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0013LO (liver_mouse)
pI: 5.89 Mw: 44157
%vol: 0.101551 %od: 0.0740745
================
*OAT_MOUSE*
accession n°: P29758
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
Spot: 2D-0013LO (liver_mouse)
pI: 5.89 Mw: 44157
%vol: 0.101551 %od: 0.0740745
================
*OAT_MOUSE*
accession n°: P29758
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
[Click to access protein entry]
Spot: 2D-0013FV (liver_mouse)
pI: 6.32 Mw: 49908
%vol: 0.147195 %od: 0.407918
================
*ALDH2_MOUSE*
accession n°: P47738
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 902.5 (0), 990.506 (0), 1040.564 (0), 1132.586 (0),
1419.742 (0), 1470.767 (0), 1506.725 (0), 1531.756 (0), 1599.772 (0),
1774.84 (0), 1844.047 (0), 2286.173 (0) }
Spot: 2D-0013FV (liver_mouse)
pI: 6.32 Mw: 49908
%vol: 0.147195 %od: 0.407918
================
*ALDH2_MOUSE*
accession n°: P47738
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 902.5 (0), 990.506 (0), 1040.564 (0), 1132.586 (0),
1419.742 (0), 1470.767 (0), 1506.725 (0), 1531.756 (0), 1599.772 (0),
1774.84 (0), 1844.047 (0), 2286.173 (0) }
[Click to access protein entry]
Spot: 2D-00140Z (liver_mouse)
pI: 6.78 Mw: 32004
%vol: 0.131375 %od: 0.0576327
================
*GLYC_MOUSE*
accession n°: P50431
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-00140Z (liver_mouse)
pI: 6.78 Mw: 32004
%vol: 0.131375 %od: 0.0576327
================
*GLYC_MOUSE*
accession n°: P50431
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0012N2 (liver_mouse)
pI: 6.31 Mw: 160000
%vol: 0.12837 %od: 0.121267
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1036.541 (0), 1090.615 (0), 1162.656 (0), 1221.675 (0),
1298.719 (0), 1322.731 (0), 1353.779 (0), 1408.707 (0), 1475.801 (0),
1484.902 (0), 1507.76 (0), 1588.835 (0), 1637.93 (0), 1711.042 (0)
}
Spot: 2D-0012N2 (liver_mouse)
pI: 6.31 Mw: 160000
%vol: 0.12837 %od: 0.121267
================
*CPSM_MOUSE*
accession n°: P99015
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1036.541 (0), 1090.615 (0), 1162.656 (0), 1221.675 (0),
1298.719 (0), 1322.731 (0), 1353.779 (0), 1408.707 (0), 1475.801 (0),
1484.902 (0), 1507.76 (0), 1588.835 (0), 1637.93 (0), 1711.042 (0)
}
[Click to access protein entry]
Spot: 2D-0012UH (liver_mouse)
pI: 4.87 Mw: 90604
%vol: 0.0979218 %od: 0.0231743
================
*ENPL_MOUSE*
accession n°: P08113
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0012UH (liver_mouse)
pI: 4.87 Mw: 90604
%vol: 0.0979218 %od: 0.0231743
================
*ENPL_MOUSE*
accession n°: P08113
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-001HE1 (liver_mouse)
pI: 6.03 Mw: 22834
%vol: 0.0102628 %od: 0.00490531
================
*GPX1_MOUSE*
accession n°: P11352
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1101.613 (0), 1136.534 (0), 1155.64 (0), 1193.602 (0),
1311.72 (0), 1957.976 (0), 2101.035 (0), 2906.382 (0)
}
Spot: 2D-001HE1 (liver_mouse)
pI: 6.03 Mw: 22834
%vol: 0.0102628 %od: 0.00490531
================
*GPX1_MOUSE*
accession n°: P11352
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1101.613 (0), 1136.534 (0), 1155.64 (0), 1193.602 (0),
1311.72 (0), 1957.976 (0), 2101.035 (0), 2906.382 (0)
}
[Click to access protein entry]
Spot: 2D-0013OX (liver_mouse)
pI: 6.43 Mw: 41076
%vol: 0.121566 %od: 0.159847
================
*IVD_MOUSE*
accession n°: P99016
Identification Methods:
* MAPPING: {Mi,Gm}
Spot: 2D-0013OX (liver_mouse)
pI: 6.43 Mw: 41076
%vol: 0.121566 %od: 0.159847
================
*IVD_MOUSE*
accession n°: P99016
Identification Methods:
* MAPPING: {Mi,Gm}
[Click to access protein entry]
Spot: 2D-0013BQ (liver_mouse)
pI: 6.72 Mw: 55784
%vol: 0.0793807 %od: 0.134238
================
*CATA_MOUSE*
accession n°: P24270
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013BQ (liver_mouse)
pI: 6.72 Mw: 55784
%vol: 0.0793807 %od: 0.134238
================
*CATA_MOUSE*
accession n°: P24270
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0014GC (liver_mouse)
pI: 4.80 Mw: 19046
%vol: 0.119468 %od: 0.172903
================
*MUP6_MOUSE*
accession n°: P02762
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 836.357 (0), 964.447 (0), 1258.646 (0), 1415.701 (0),
1502.701 (0), 1839.879 (0), 1895.116 (0) }
================
*MUP8_MOUSE*
accession n: P04938
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.529 (0), 1258.679 (0), 1415.741 (0), 1502.696 (0),
1528.799 (0), 1596.786 (0), 1839.813 (0), 1895.09 (0), 2009.912 (0)
}
Spot: 2D-0014GC (liver_mouse)
pI: 4.80 Mw: 19046
%vol: 0.119468 %od: 0.172903
================
*MUP6_MOUSE*
accession n°: P02762
Identification Methods:
* MAPPING: {Mi,Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 836.357 (0), 964.447 (0), 1258.646 (0), 1415.701 (0),
1502.701 (0), 1839.879 (0), 1895.116 (0) }
================
*MUP8_MOUSE*
accession n: P04938
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.529 (0), 1258.679 (0), 1415.741 (0), 1502.696 (0),
1528.799 (0), 1596.786 (0), 1839.813 (0), 1895.09 (0), 2009.912 (0)
}
[Click to access protein entry]
Spot: 2D-0014KD (liver_mouse)
pI: 6.56 Mw: 13235
%vol: 0.0484223 %od: 0.11196
================
*NDKA_MOUSE*
accession n°: P15532
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1051.488 (0), 1163.659 (0), 1179.651 (0), 1344.766 (0),
1909.94 (0) }
Spot: 2D-0014KD (liver_mouse)
pI: 6.56 Mw: 13235
%vol: 0.0484223 %od: 0.11196
================
*NDKA_MOUSE*
accession n°: P15532
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1051.488 (0), 1163.659 (0), 1179.651 (0), 1344.766 (0),
1909.94 (0) }
[Click to access protein entry]
Spot: 2D-0013U2 (liver_mouse)
pI: 6.67 Mw: 37081
%vol: 0.151901 %od: 0.174169
================
*ARGI1_MOUSE*
accession n°: Q61176
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 911.402 (0), 948.542 (0), 1152.585 (0), 1161.62 (0),
1285.636 (0), 1357.672 (0), 1444.718 (0), 1680.882 (0), 1873.989 (0),
2018.091 (0), 2203.097 (0), 3269.793 (0) }
Spot: 2D-0013U2 (liver_mouse)
pI: 6.67 Mw: 37081
%vol: 0.151901 %od: 0.174169
================
*ARGI1_MOUSE*
accession n°: Q61176
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 911.402 (0), 948.542 (0), 1152.585 (0), 1161.62 (0),
1285.636 (0), 1357.672 (0), 1444.718 (0), 1680.882 (0), 1873.989 (0),
2018.091 (0), 2203.097 (0), 3269.793 (0) }
[Click to access protein entry]
Spot: 2D-0014B6 (liver_mouse)
pI: 8.40 Mw: 25232
%vol: 0.0570407 %od: 0.0443459
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0014B6 (liver_mouse)
pI: 8.40 Mw: 25232
%vol: 0.0570407 %od: 0.0443459
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-00131T (liver_mouse)
pI: 5.28 Mw: 71490
%vol: 0.0463811 %od: 0.0198552
================
*NCPR_MOUSE*
accession n°: P37040
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-00131T (liver_mouse)
pI: 5.28 Mw: 71490
%vol: 0.0463811 %od: 0.0198552
================
*NCPR_MOUSE*
accession n°: P37040
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0014IO (liver_mouse)
pI: 4.08 Mw: 14659
%vol: 0.0151957 %od: 0.0434171
================
*CALM_MOUSE*
accession n°: P62204
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0014IO (liver_mouse)
pI: 4.08 Mw: 14659
%vol: 0.0151957 %od: 0.0434171
================
*CALM_MOUSE*
accession n°: P62204
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0014BA (liver_mouse)
pI: 8.26 Mw: 25179
%vol: 0.103592 %od: 0.096662
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0014BA (liver_mouse)
pI: 8.26 Mw: 25179
%vol: 0.103592 %od: 0.096662
================
*GSTP1_MOUSE*
accession n°: P19157
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013DM (liver_mouse)
pI: 5.11 Mw: 53504
%vol: 0.0784735 %od: 0.0497805
================
*TBA1A_MOUSE*
accession n°: P68369
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1085.646 (0), 1410.75 (0), 1701.918 (0), 2409.167 (0)
}
Spot: 2D-0013DM (liver_mouse)
pI: 5.11 Mw: 53504
%vol: 0.0784735 %od: 0.0497805
================
*TBA1A_MOUSE*
accession n°: P68369
Identification Methods:
* MAPPING: {PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 1085.646 (0), 1410.75 (0), 1701.918 (0), 2409.167 (0)
}
[Click to access protein entry]
Spot: 2D-0013FP (liver_mouse)
pI: 6.16 Mw: 50467
%vol: 0.121169 %od: 0.104618
================
*ALDH2_MOUSE*
accession n°: P47738
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013FP (liver_mouse)
pI: 6.16 Mw: 50467
%vol: 0.121169 %od: 0.104618
================
*ALDH2_MOUSE*
accession n°: P47738
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013DW (liver_mouse)
pI: 5.46 Mw: 53207
%vol: 0.0946899 %od: 0.126683
================
*SBP1_MOUSE*
accession n°: P17563
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013DW (liver_mouse)
pI: 5.46 Mw: 53207
%vol: 0.0946899 %od: 0.126683
================
*SBP1_MOUSE*
accession n°: P17563
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0012V6 (liver_mouse)
pI: 6.84 Mw: 89040
%vol: 0.0489893 %od: 0.0256466
================
*ACOC_MOUSE*
accession n°: P28271
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1458.762 (0), 1599.822 (0), 1852.962 (0), 2060.166 (0),
2065.103 (0), 2090.172 (0), 2381.178 (0) }
Spot: 2D-0012V6 (liver_mouse)
pI: 6.84 Mw: 89040
%vol: 0.0489893 %od: 0.0256466
================
*ACOC_MOUSE*
accession n°: P28271
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1458.762 (0), 1599.822 (0), 1852.962 (0), 2060.166 (0),
2065.103 (0), 2090.172 (0), 2381.178 (0) }
[Click to access protein entry]
Spot: 2D-00145G (liver_mouse)
pI: 6.98 Mw: 28122
%vol: 0.171859 %od: 1.47997
================
*CAH3_MOUSE*
accession n°: P16015
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-00145G (liver_mouse)
pI: 6.98 Mw: 28122
%vol: 0.171859 %od: 1.47997
================
*CAH3_MOUSE*
accession n°: P16015
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-00132W (liver_mouse)
pI: 5.30 Mw: 69426
%vol: 0.0538088 %od: 0.0440283
================
*GRP75_MOUSE*
accession n°: P38647
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
Spot: 2D-00132W (liver_mouse)
pI: 5.30 Mw: 69426
%vol: 0.0538088 %od: 0.0440283
================
*GRP75_MOUSE*
accession n°: P38647
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
[Click to access protein entry]
Spot: 2D-0013UU (liver_mouse)
pI: 6.37 Mw: 36671
%vol: 0.0853343 %od: 0.0721506
================
*HEM2_MOUSE*
accession n°: P10518
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1090.582 (0), 1194.533 (0), 1221.559 (0), 1251.542 (0),
1255.63 (0), 2203.191 (0), 2539.237 (0) }
Spot: 2D-0013UU (liver_mouse)
pI: 6.37 Mw: 36671
%vol: 0.0853343 %od: 0.0721506
================
*HEM2_MOUSE*
accession n°: P10518
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1090.582 (0), 1194.533 (0), 1221.559 (0), 1251.542 (0),
1255.63 (0), 2203.191 (0), 2539.237 (0) }
[Click to access protein entry]
Spot: 2D-001485 (liver_mouse)
pI: 8.07 Mw: 27086
%vol: 0.136705 %od: 0.606674
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1105.523 (0), 1248.672 (0), 1264.6 (0), 1898.897 (0),
1993.89 (0) }
Spot: 2D-001485 (liver_mouse)
pI: 8.07 Mw: 27086
%vol: 0.136705 %od: 0.606674
================
*GSTM1_MOUSE*
accession n°: P10649
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1105.523 (0), 1248.672 (0), 1264.6 (0), 1898.897 (0),
1993.89 (0) }
[Click to access protein entry]
Spot: 2D-0013YI (liver_mouse)
pI: 7.51 Mw: 33718
%vol: 0.0810818 %od: 0.0619371
================
*THTR_MOUSE*
accession n°: P52196
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 940.424 (0), 1049.585 (0), 1351.642 (0), 1663.723 (0),
2066.996 (0) }
Spot: 2D-0013YI (liver_mouse)
pI: 7.51 Mw: 33718
%vol: 0.0810818 %od: 0.0619371
================
*THTR_MOUSE*
accession n°: P52196
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 940.424 (0), 1049.585 (0), 1351.642 (0), 1663.723 (0),
2066.996 (0) }
[Click to access protein entry]
Spot: 2D-0013CZ (liver_mouse)
pI: 5.73 Mw: 54556
%vol: 0.124968 %od: 0.0960781
================
*KTRB_MOUSE*
accession n°: P99019
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013CZ (liver_mouse)
pI: 5.73 Mw: 54556
%vol: 0.124968 %od: 0.0960781
================
*KTRB_MOUSE*
accession n°: P99019
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-001344 (liver_mouse)
pI: 5.80 Mw: 66529
%vol: 0.125365 %od: 0.133178
================
*ALBU_MOUSE*
accession n°: P07724
Identification Methods:
* MAPPING: {Mi,Gm}
Spot: 2D-001344 (liver_mouse)
pI: 5.80 Mw: 66529
%vol: 0.125365 %od: 0.133178
================
*ALBU_MOUSE*
accession n°: P07724
Identification Methods:
* MAPPING: {Mi,Gm}
[Click to access protein entry]
Spot: 2D-0012WZ (liver_mouse)
pI: 6.54 Mw: 82896
%vol: 0.0957105 %od: 0.0588365
================
*M2GD_MOUSE*
accession n°: P99039
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 932.541 (0), 973.533 (0), 1104.581 (0), 1115.562 (0),
1123.553 (0), 1131.658 (0), 1482.702 (0), 1487.74 (0), 1723.82 (0),
1983.011 (0) }
Spot: 2D-0012WZ (liver_mouse)
pI: 6.54 Mw: 82896
%vol: 0.0957105 %od: 0.0588365
================
*M2GD_MOUSE*
accession n°: P99039
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 932.541 (0), 973.533 (0), 1104.581 (0), 1115.562 (0),
1123.553 (0), 1131.658 (0), 1482.702 (0), 1487.74 (0), 1723.82 (0),
1983.011 (0) }
[Click to access protein entry]
Spot: 2D-0014LT (liver_mouse)
pI: 7.13 Mw: 12075
%vol: 0.08936 %od: 0.219168
================
*B2MG_MOUSE*
accession n°: P01887
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014LT (liver_mouse)
pI: 7.13 Mw: 12075
%vol: 0.08936 %od: 0.219168
================
*B2MG_MOUSE*
accession n°: P01887
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0013GJ (liver_mouse)
pI: 4.97 Mw: 49770
%vol: 0.065319 %od: 0.0534036
================
*TBB5_MOUSE*
accession n°: P99024
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0013GJ (liver_mouse)
pI: 4.97 Mw: 49770
%vol: 0.065319 %od: 0.0534036
================
*TBB5_MOUSE*
accession n°: P99024
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0013EH (liver_mouse)
pI: 5.92 Mw: 52472
%vol: 0.118447 %od: 0.111989
================
*SBP1_MOUSE*
accession n°: P17563
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1051.54 (0), 1068.669 (0), 1149.577 (0), 1214.618 (0),
1233.643 (0), 1330.779 (0), 1345.7 (0), 1454.734 (0), 1667.81 (0),
1689.84 (0), 1707.812 (0), 2283.275 (0) }
Spot: 2D-0013EH (liver_mouse)
pI: 5.92 Mw: 52472
%vol: 0.118447 %od: 0.111989
================
*SBP1_MOUSE*
accession n°: P17563
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1051.54 (0), 1068.669 (0), 1149.577 (0), 1214.618 (0),
1233.643 (0), 1330.779 (0), 1345.7 (0), 1454.734 (0), 1667.81 (0),
1689.84 (0), 1707.812 (0), 2283.275 (0) }
[Click to access protein entry]
Spot: 2D-0013BT (liver_mouse)
pI: 4.86 Mw: 55474
%vol: 0.146798 %od: 0.420623
================
*PDIA1_MOUSE*
accession n°: P09103
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 905.318 (0), 910.361 (0), 966.512 (0), 1051.491 (0), 1066.5 (0)
}
Spot: 2D-0013BT (liver_mouse)
pI: 4.86 Mw: 55474
%vol: 0.146798 %od: 0.420623
================
*PDIA1_MOUSE*
accession n°: P09103
Identification Methods:
* MAPPING: {Mi,PMF,Gm}
peptide masses: { (TRYPSIN)
$m/Z= 905.318 (0), 910.361 (0), 966.512 (0), 1051.491 (0), 1066.5 (0)
}
[Click to access protein entry]
Spot: 2D-00132M (liver_mouse)
pI: 5.04 Mw: 69426
%vol: 0.138803 %od: 0.431864
================
*GRP78_MOUSE*
accession n°: P20029
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 986.493 (0), 1074.527 (0), 1191.599 (0), 1228.626 (0),
1233.622 (0), 1329.623 (0), 1430.69 (0), 1460.751 (0), 1566.765 (0),
1604.867 (0), 1659.916 (0), 1677.833 (0), 1836.955 (0), 1887.952 (0),
1933.97 (0), 1999.061 (0), 2148.947 (0) }
Spot: 2D-00132M (liver_mouse)
pI: 5.04 Mw: 69426
%vol: 0.138803 %od: 0.431864
================
*GRP78_MOUSE*
accession n°: P20029
Identification Methods:
* MAPPING: {Gm} {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 986.493 (0), 1074.527 (0), 1191.599 (0), 1228.626 (0),
1233.622 (0), 1329.623 (0), 1430.69 (0), 1460.751 (0), 1566.765 (0),
1604.867 (0), 1659.916 (0), 1677.833 (0), 1836.955 (0), 1887.952 (0),
1933.97 (0), 1999.061 (0), 2148.947 (0) }
[Click to access protein entry]
Spot: 2D-0014IK (liver_mouse)
pI: 4.68 Mw: 14999
%vol: 0.0125308 %od: 0.0093838
================
*CYB5_MOUSE*
accession n°: P56395
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1120.632 (0), 1428.802 (0), 1511.838 (0), 2206.025 (0)
}
Spot: 2D-0014IK (liver_mouse)
pI: 4.68 Mw: 14999
%vol: 0.0125308 %od: 0.0093838
================
*CYB5_MOUSE*
accession n°: P56395
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1120.632 (0), 1428.802 (0), 1511.838 (0), 2206.025 (0)
}
[Click to access protein entry]
Spot: 2D-0014GI (liver_mouse)
pI: 4.74 Mw: 18844
%vol: 0.0550561 %od: 0.0695042
================
*MUP6_MOUSE*
accession n°: P02762
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.41 (0), 1221.538 (0), 1258.577 (0), 1415.724 (0),
1502.687 (0), 1596.862 (0), 1839.88 (0), 1895.058 (0), 2009.984 (0)
}
================
*MUP8_MOUSE*
accession n: P04938
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.41 (0), 1221.538 (0), 1258.577 (0), 1415.724 (0),
1502.687 (0), 1596.862 (0), 1839.88 (0), 1895.058 (0), 2009.984 (0)
}
Spot: 2D-0014GI (liver_mouse)
pI: 4.74 Mw: 18844
%vol: 0.0550561 %od: 0.0695042
================
*MUP6_MOUSE*
accession n°: P02762
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.41 (0), 1221.538 (0), 1258.577 (0), 1415.724 (0),
1502.687 (0), 1596.862 (0), 1839.88 (0), 1895.058 (0), 2009.984 (0)
}
================
*MUP8_MOUSE*
accession n: P04938
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 964.41 (0), 1221.538 (0), 1258.577 (0), 1415.724 (0),
1502.687 (0), 1596.862 (0), 1839.88 (0), 1895.058 (0), 2009.984 (0)
}
[Click to access protein entry]
Spot: 2D-00135L (liver_mouse)
pI: 6.85 Mw: 65185
%vol: 0.0727468 %od: 0.0594067
================
*TKT_MOUSE*
accession n°: P40142
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 869.566 (0), 978.556 (0), 1264.646 (0), 1402.741 (0),
1413.805 (0), 2020.157 (0), 2481.184 (0) }
Spot: 2D-00135L (liver_mouse)
pI: 6.85 Mw: 65185
%vol: 0.0727468 %od: 0.0594067
================
*TKT_MOUSE*
accession n°: P40142
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 869.566 (0), 978.556 (0), 1264.646 (0), 1402.741 (0),
1413.805 (0), 2020.157 (0), 2481.184 (0) }
[Click to access protein entry]
Spot: 2D-0013XS (liver_mouse)
pI: 5.01 Mw: 34152
%vol: 0.0405408 %od: 0.0532243
================
*RGN_MOUSE*
accession n°: Q64374
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1057.556 (0), 1138.549 (0), 1195.548 (0), 1207.657 (0),
1232.588 (0), 1283.735 (0), 1410.593 (0), 1714.874 (0), 1871.898 (0),
3207.525 (0) }
Spot: 2D-0013XS (liver_mouse)
pI: 5.01 Mw: 34152
%vol: 0.0405408 %od: 0.0532243
================
*RGN_MOUSE*
accession n°: Q64374
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1057.556 (0), 1138.549 (0), 1195.548 (0), 1207.657 (0),
1232.588 (0), 1283.735 (0), 1410.593 (0), 1714.874 (0), 1871.898 (0),
3207.525 (0) }
[Click to access protein entry]
Spot: 2D-0013XY (liver_mouse)
pI: 4.97 Mw: 34076
%vol: 0.0318658 %od: 0.0239717
================
*RGN_MOUSE*
accession n°: Q64374
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1057.565 (0), 1232.607 (0), 1283.757 (0), 1714.861 (0),
1871.886 (0) }
Spot: 2D-0013XY (liver_mouse)
pI: 4.97 Mw: 34076
%vol: 0.0318658 %od: 0.0239717
================
*RGN_MOUSE*
accession n°: Q64374
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 1057.565 (0), 1232.607 (0), 1283.757 (0), 1714.861 (0),
1871.886 (0) }
[Click to access protein entry]
Spot: 2D-0013DX (liver_mouse)
pI: 5.54 Mw: 53207
%vol: 0.0555098 %od: 0.0432719
================
*K2C1_MOUSE*
accession n°: P04104
Identification Methods:
* MAPPING: {Gm}
Spot: 2D-0013DX (liver_mouse)
pI: 5.54 Mw: 53207
%vol: 0.0555098 %od: 0.0432719
================
*K2C1_MOUSE*
accession n°: P04104
Identification Methods:
* MAPPING: {Gm}
[Click to access protein entry]
Spot: 2D-0013EE (liver_mouse)
pI: 7.83 Mw: 52764
%vol: 0.0729736 %od: 0.0717632
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
Spot: 2D-0013EE (liver_mouse)
pI: 7.83 Mw: 52764
%vol: 0.0729736 %od: 0.0717632
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
[Click to access protein entry]
Spot: 2D-0013EK (liver_mouse)
pI: 7.49 Mw: 52180
%vol: 0.141525 %od: 0.309844
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
================
*HMCS2_MOUSE*
accession n: P54869
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 994.55 (0), 1052.584 (0), 1095.668 (0), 1142.594 (0),
1627.67 (0), 1643.695 (0), 1884.948 (0), 2299.146 (0), 2397.17 (0),
2626.288 (0) }
Spot: 2D-0013EK (liver_mouse)
pI: 7.49 Mw: 52180
%vol: 0.141525 %od: 0.309844
================
*AL1A1_MOUSE*
accession n°: P24549
Identification Methods:
* MAPPING: {PMF}
================
*HMCS2_MOUSE*
accession n: P54869
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 994.55 (0), 1052.584 (0), 1095.668 (0), 1142.594 (0),
1627.67 (0), 1643.695 (0), 1884.948 (0), 2299.146 (0), 2397.17 (0),
2626.288 (0) }
[Click to access protein entry]
Spot: 2D-0014NC (liver_mouse)
pI: 6.32 Mw: 10149
%vol: 0.0441131 %od: 0.0518311
================
*GFRP_MOUSE*
accession n°: P99025
Identification Methods:
* MAPPING: {Mi}
Spot: 2D-0014NC (liver_mouse)
pI: 6.32 Mw: 10149
%vol: 0.0441131 %od: 0.0518311
================
*GFRP_MOUSE*
accession n°: P99025
Identification Methods:
* MAPPING: {Mi}
[Click to access protein entry]
Spot: 2D-0013MM (liver_mouse)
pI: 5.88 Mw: 43185
%vol: 0.0376492 %od: 0.00900133
================
*OAT_MOUSE*
accession n°: P29758
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 909.561 (0), 968.661 (0), 1241.58 (0), 1492.889 (0),
1639.762 (0), 1736.905 (0), 1810.968 (0), 1952.888 (0), 2108.038 (0),
2312.028 (0) }
Spot: 2D-0013MM (liver_mouse)
pI: 5.88 Mw: 43185
%vol: 0.0376492 %od: 0.00900133
================
*OAT_MOUSE*
accession n°: P29758
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 909.561 (0), 968.661 (0), 1241.58 (0), 1492.889 (0),
1639.762 (0), 1736.905 (0), 1810.968 (0), 1952.888 (0), 2108.038 (0),
2312.028 (0) }
[Click to access protein entry]
Spot: 2D-0013ZZ (liver_mouse)
pI: 6.83 Mw: 32611
%vol: 0.14589 %od: 0.291922
================
*GLMT_MOUSE*
accession n°: P99051
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 882.385 (0), 941.533 (0), 1250.626 (0), 1864.014 (0),
1989.99 (0) }
Spot: 2D-0013ZZ (liver_mouse)
pI: 6.83 Mw: 32611
%vol: 0.14589 %od: 0.291922
================
*GLMT_MOUSE*
accession n°: P99051
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 882.385 (0), 941.533 (0), 1250.626 (0), 1864.014 (0),
1989.99 (0) }
[Click to access protein entry]
Spot: 2D-0013FA (liver_mouse)
pI: 6.69 Mw: 51459
%vol: 0.111417 %od: 0.29352
================
*DHE3_MOUSE*
accession n°: P26443
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.539 (0), 1000.48 (0), 1016.467 (0), 1059.549 (0),
1196.677 (0), 1425.645 (0), 1491.717 (0), 1507.722 (0), 1581.74 (0),
1711.782 (0), 1723.893 (0), 1748.913 (0), 1764.896 (0), 1894.09 (0),
1920.959 (0), 1931.916 (0), 1936.907 (0), 2060.021 (0), 2242.201 (0)
}
Spot: 2D-0013FA (liver_mouse)
pI: 6.69 Mw: 51459
%vol: 0.111417 %od: 0.29352
================
*DHE3_MOUSE*
accession n°: P26443
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 963.539 (0), 1000.48 (0), 1016.467 (0), 1059.549 (0),
1196.677 (0), 1425.645 (0), 1491.717 (0), 1507.722 (0), 1581.74 (0),
1711.782 (0), 1723.893 (0), 1748.913 (0), 1764.896 (0), 1894.09 (0),
1920.959 (0), 1931.916 (0), 1936.907 (0), 2060.021 (0), 2242.201 (0)
}
SWISS-2DPAGE Viewer
|
Back to the |
-Get more information by dragging your mouse pointer over any highlighted spot -Click on a highlighted spot to access all its associated protein entries
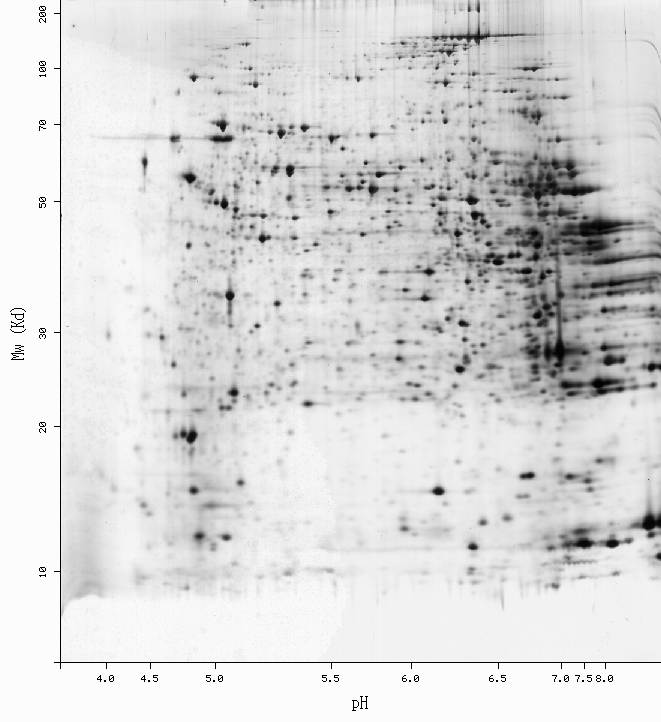
Back to the