Attention: World-2DPAGE is no longer maintained.
[Click to access protein entry]
Spot: 2D-001LQC (ecoli5-6)
pI: 5.78 Mw: 17897
%vol: 0.179311 %od: 0.212426
================
*DPS_ECOLI*
accession n°: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 942.642 (0), 951.751 (0), 1518.247 (0), 1677.213 (0),
1679.206 (0), 1693.222 (0) }
Spot: 2D-001LQC (ecoli5-6)
pI: 5.78 Mw: 17897
%vol: 0.179311 %od: 0.212426
================
*DPS_ECOLI*
accession n°: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 942.642 (0), 951.751 (0), 1518.247 (0), 1677.213 (0),
1679.206 (0), 1693.222 (0) }
[Click to access protein entry]
Spot: 2D-001LR4 (ecoli5-6)
pI: 5.27 Mw: 16787
%vol: 0.11773 %od: 0.137014
================
*DPS_ECOLI*
accession n°: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 915.56 (0), 951.536 (0), 1020.57 (0), 1043.61 (0), 1501.652 (0),
1517.949 (0), 1678.831 (0), 1692.796 (0) }
Spot: 2D-001LR4 (ecoli5-6)
pI: 5.27 Mw: 16787
%vol: 0.11773 %od: 0.137014
================
*DPS_ECOLI*
accession n°: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 915.56 (0), 951.536 (0), 1020.57 (0), 1043.61 (0), 1501.652 (0),
1517.949 (0), 1678.831 (0), 1692.796 (0) }
[Click to access protein entry]
Spot: 2D-001LRH (ecoli5-6)
pI: 5.21 Mw: 16164
%vol: 0.133168 %od: 0.246888
================
*DPS_ECOLI*
accession n°: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 951.14 (0), 1020.12 (0), 1501.09 (0), 1517.204 (0),
1678.142 (0), 1692.113 (0) }
Spot: 2D-001LRH (ecoli5-6)
pI: 5.21 Mw: 16164
%vol: 0.133168 %od: 0.246888
================
*DPS_ECOLI*
accession n°: P0ABT2
Identification Methods:
* MAPPING: {PMF}
peptide masses: { (TRYPSIN)
$m/Z= 951.14 (0), 1020.12 (0), 1501.09 (0), 1517.204 (0),
1678.142 (0), 1692.113 (0) }
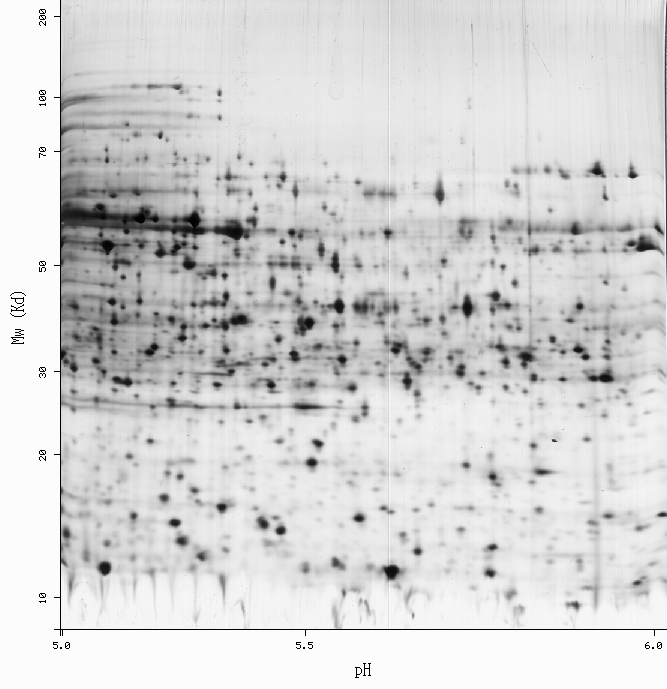
SWISS-2DPAGE Viewer
|
Back to the |
-Get more information by dragging your mouse pointer over any highlighted spot -Click on a highlighted spot to access all its associated protein entries
[estimated location]
Back to the