| General information about the entry |
| View entry in simple text format |
| Entry name | NPM_MOUSE |
| Primary accession number | Q61937 |
| integrated into World-2DPAGE Repository (0021) on | June 9, 2010 (release 1) |
| 2D Annotations were last modified on | June 20, 2011 (version 2) |
| General Annotations were last modified on | November 30, 2011 (version 2) |
| Name and origin of the protein |
| Description | RecName: Full=Nucleophosmin; Short=NPM; AltName: Full=Nucleolar phosphoprotein B23; AltName: Full=Nucleolar protein NO38; AltName: Full=Numatrin;. |
| Gene name | Name=Npm1
|
| Annotated species | Mus musculus (Mouse) [TaxID: 10090] |
| Taxonomy | Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; Eutheria; Euarchontoglires; Glires; Rodentia; Sciurognathi; Muroidea; Muridae; Murinae; Mus; Mus. |
| References |
| [1] |
2D-PAGE GEL CHARACTERIZATION
De Cegli R., Romito A., Iacobacci S., Mao L., Lauria M., Fedele A.O., Klose J., Borel C., Descombes P., Antonarakis S.E., Di Bernardo D., Banfi S., Ballabio A., Cobellis G.
''A mouse Embryonic Stem Cell Bank for inducible overexpression of human Chromosome 21 genes''
Genome Biol 11(6):R64 (2010)
|
|
| 2D PAGE maps for identified proteins
|
|
How to interpret a protein
|
RUNX1_DAY7 {Mouse embryonic stem cells overexpressing Runx1, Day 7}
Mus musculus (Mouse)
Tissue: Embryonic stem cell
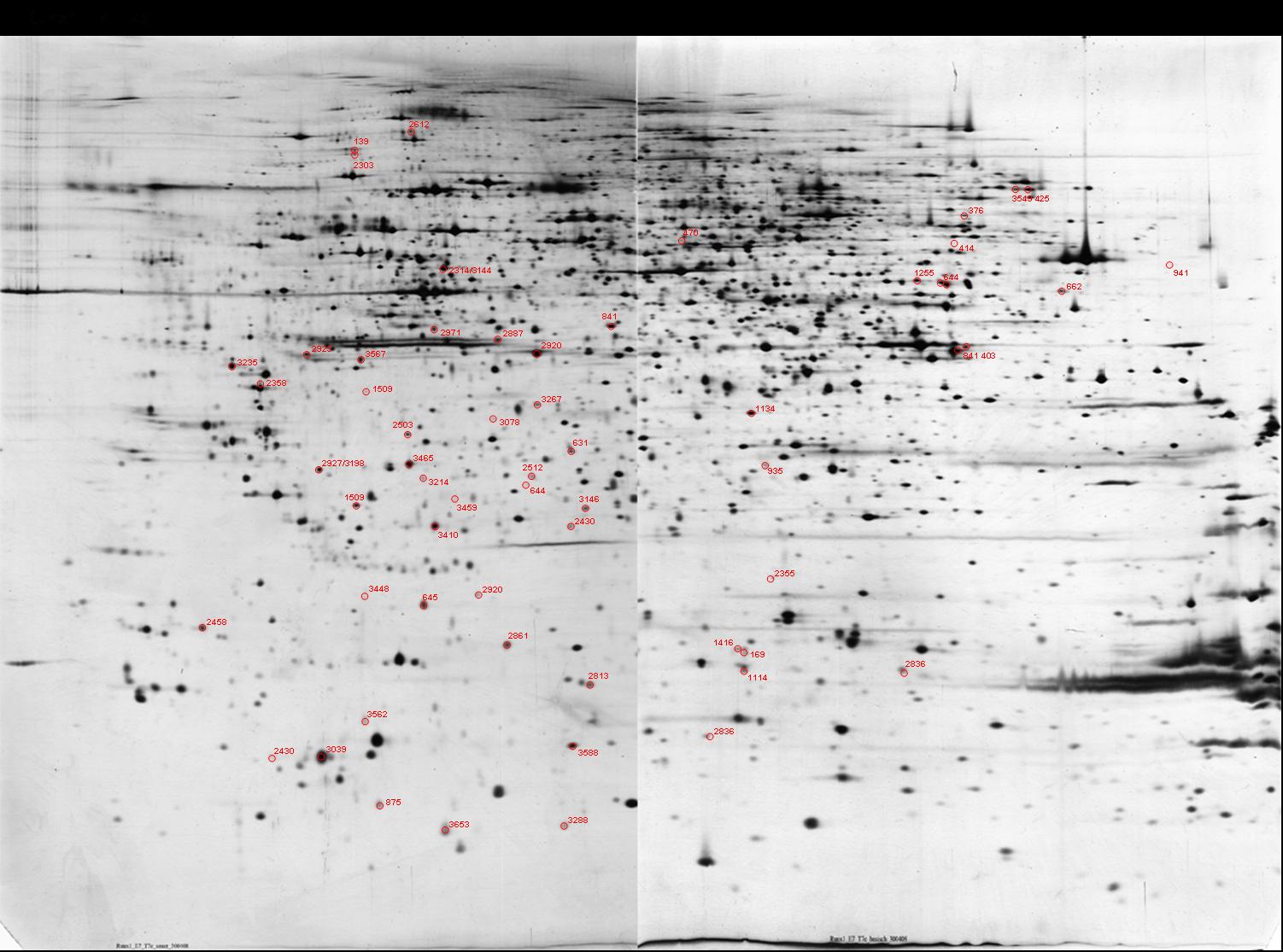
map experimental info
|
|
RUNX1_DAY7
MAP LOCATIONS:
EXPRESSION:
| SPOT 2923: Protein ratio 48h/0h=1.28 [1]. |
IDENTIFICATION: SPOT 2923: Mascot Protein score=242. SeqCov=10%. Peptides identified=6 [1].
MAPPING (identification):
|
| Copyright |
| Data from Dr. Lei Mao, Institute of Human Genetics, Charite Universitaetsmedizin Berlin, Germany |
| Cross-references |
| UniProtKB/Swiss-Prot | Q61937; NPM_MOUSE. |