|
|
|
|
|
Protein list for SWISS-2DPAGE ECOLI 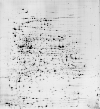
Escherichia coli
|
Sample Preparation / Gel Related Documents
Protein list for ecoli 206 identified spots / 180 protein entries
| Gene Name | Description | Accession n° | Spot ID | Exp pI | Exp Mw | Mapping Methods | Comment Topics | References |
|---|
| accB | RecName: Full=Biotin carboxyl carrier protein of acetyl-CoA carboxylase; Short=BCCP; | P0ABD8
(BCCP_ECOLI) |
|
|
|
|
|
|
| aceE | RecName: Full=Pyruvate dehydrogenase E1 component; EC=1.2.4.1; | P0AFG8
(ODP1_ECOLI) |
|
|
|
|
|
|
| aceF | RecName: Full=Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex; EC=2.3.1.12; AltName: Full=Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex; AltName: Full=E2; | P06959
(ODP2_ECOLI) |
|
|
|
|
|
|
| ackA | RecName: Full=Acetate kinase; EC=2.7.2.1; AltName: Full=Acetokinase; | P0A6A3
(ACKA_ECOLI) |
|
|
|
|
|
|
| adk | RecName: Full=Adenylate kinase; Short=AK; EC=2.7.4.3; AltName: Full=ATP-AMP transphosphorylase; | P69441
(KAD_ECOLI) |
|
|
|
|
|
|
| ahpC | RecName: Full=Alkyl hydroperoxide reductase subunit C; EC=1.11.1.15; AltName: Full=Alkyl hydroperoxide reductase protein C22; AltName: Full=Peroxiredoxin; AltName: Full=SCRP-23; AltName: Full=Sulfate starvation-induced protein 8; Short=SSI8; AltName: Full=Thioredoxin peroxidase; | P0AE08
(AHPC_ECOLI) |
|
|
|
|
|
|
| alaS | RecName: Full=Alanyl-tRNA synthetase; EC=6.1.1.7; AltName: Full=Alanine--tRNA ligase; Short=AlaRS; | P00957
(SYA_ECOLI) |
|
|
|
|
|
|
| appA | RecName: Full=Periplasmic AppA protein; Includes: RecName: Full=Phosphoanhydride phosphohydrolase; EC=3.1.3.2; AltName: Full=pH 2.5 acid phosphatase; Short=AP; Includes: RecName: Full=4-phytase; EC=3.1.3.26; Flags: Precursor; | P07102
(PPA_ECOLI) |
|
|
|
|
|
|
| argI | RecName: Full=Ornithine carbamoyltransferase chain I; EC=2.1.3.3; AltName: Full=OTCase-1; | P04391
(OTC1_ECOLI) |
|
|
|
|
|
|
| argS | RecName: Full=Arginyl-tRNA synthetase; EC=6.1.1.19; AltName: Full=Arginine--tRNA ligase; Short=ArgRS; | P11875
(SYR_ECOLI) |
|
|
|
|
|
|
| aroG | RecName: Full=Phospho-2-dehydro-3-deoxyheptonate aldolase, Phe-sensitive; EC=2.5.1.54; AltName: Full=3-deoxy-D-arabino-heptulosonate 7-phosphate synthase; AltName: Full=DAHP synthase; AltName: Full=Phospho-2-keto-3-deoxyheptonate aldolase; | P0AB91
(AROG_ECOLI) |
|
|
|
|
|
|
| aroK | RecName: Full=Shikimate kinase 1; Short=SK 1; EC=2.7.1.71; AltName: Full=Shikimate kinase I; Short=SKI; | P0A6D7
(AROK_ECOLI) |
|
|
|
|
|
|
| artI | RecName: Full=Putative ABC transporter arginine-binding protein 2; Flags: Precursor; | P30859
(ARTI_ECOLI) |
|
|
|
|
|
|
| asnS | RecName: Full=Asparaginyl-tRNA synthetase; EC=6.1.1.22; AltName: Full=Asparagine--tRNA ligase; Short=AsnRS; | P0A8M0
(SYN_ECOLI) |
|
|
|
|
|
|
| aspC | RecName: Full=Aspartate aminotransferase; Short=AspAT; EC=2.6.1.1; AltName: Full=Transaminase A; | P00509
(AAT_ECOLI) |
|
|
|
|
|
|
| aspS | RecName: Full=Aspartyl-tRNA synthetase; EC=6.1.1.12; AltName: Full=Aspartate--tRNA ligase; Short=AspRS; | P21889
(SYD_ECOLI) |
|
|
|
|
|
|
| atpA | RecName: Full=ATP synthase subunit alpha; EC=3.6.3.14; AltName: Full=ATP synthase F1 sector subunit alpha; AltName: Full=F-ATPase subunit alpha; | P0ABB0
(ATPA_ECOLI) |
|
|
|
|
|
|
| atpC | RecName: Full=ATP synthase epsilon chain; AltName: Full=ATP synthase F1 sector epsilon subunit; AltName: Full=F-ATPase epsilon subunit; | P0A6E6
(ATPE_ECOLI) |
|
|
|
|
|
|
| atpD | RecName: Full=ATP synthase subunit beta; EC=3.6.3.14; AltName: Full=ATP synthase F1 sector subunit beta; AltName: Full=F-ATPase subunit beta; | P0ABB4
(ATPB_ECOLI) |
|
|
|
|
|
|
| bcp | RecName: Full=Putative peroxiredoxin bcp; EC=1.11.1.15; AltName: Full=Bacterioferritin comigratory protein; AltName: Full=Thioredoxin reductase; | P0AE52
(BCP_ECOLI) |
|
|
|
|
|
|
| carA | RecName: Full=Carbamoyl-phosphate synthase small chain; EC=6.3.5.5; AltName: Full=Carbamoyl-phosphate synthetase glutamine chain; | P0A6F1
(CARA_ECOLI) |
|
|
|
|
|
|
| cheZ | RecName: Full=Protein phosphatase CheZ; EC=3.1.3.-; AltName: Full=Chemotaxis protein CheZ; | P0A9H9
(CHEZ_ECOLI) |
|
|
|
|
|
|
| clpB | RecName: Full=Chaperone protein ClpB; AltName: Full=Heat shock protein F84.1; | P63284
(CLPB_ECOLI) |
|
|
|
|
|
|
| crr | RecName: Full=Glucose-specific phosphotransferase enzyme IIA component; EC=2.7.1.-; AltName: Full=EIIA-Glc; AltName: Full=EIII-Glc; AltName: Full=PTS system glucose-specific EIIA component; | P69783
(PTGA_ECOLI) |
|
|
|
|
|
|
| cspC | RecName: Full=Cold shock-like protein CspC; Short=CSP-C; | P0A9Y6
(CSPC_ECOLI) |
|
|
|
|
|
|
| cysK | RecName: Full=Cysteine synthase A; Short=CSase A; EC=2.5.1.47; AltName: Full=O-acetylserine (thiol)-lyase A; Short=OAS-TL A; AltName: Full=O-acetylserine sulfhydrylase A; AltName: Full=Sulfate starvation-induced protein 5; Short=SSI5; | P0ABK5
(CYSK_ECOLI) |
|
|
|
|
|
|
| dapA | RecName: Full=Dihydrodipicolinate synthase; Short=DHDPS; EC=4.2.1.52; | P0A6L2
(DAPA_ECOLI) |
|
|
|
|
|
|
| dapD | RecName: Full=2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase; EC=2.3.1.117; AltName: Full=Tetrahydrodipicolinate N-succinyltransferase; Short=THDP succinyltransferase; Short=THP succinyltransferase; Short=Tetrahydropicolinate succinylase; | P0A9D8
(DAPD_ECOLI) |
|
|
|
|
|
|
| degQ | RecName: Full=Protease degQ; EC=3.4.21.-; Flags: Precursor; | P39099
(DEGQ_ECOLI) |
|
|
|
|
|
|
| dksA | RecName: Full=DnaK suppressor protein; | P0ABS1
(DKSA_ECOLI) |
|
|
|
|
|
|
| dnaB | RecName: Full=Replicative DNA helicase; EC=3.6.4.12; | P0ACB0
(DNAB_ECOLI) |
|
|
|
|
|
|
| dnaK | RecName: Full=Chaperone protein DnaK; AltName: Full=HSP70; AltName: Full=Heat shock 70 kDa protein; AltName: Full=Heat shock protein 70; | P0A6Y8
(DNAK_ECOLI) |
|
|
|
|
|
|
| dnaQ | RecName: Full=DNA polymerase III subunit epsilon; EC=2.7.7.7; | P03007
(DPO3E_ECOLI) |
|
|
|
|
|
|
| dppA | RecName: Full=Periplasmic dipeptide transport protein; AltName: Full=Dipeptide-binding protein; Short=DBP; Flags: Precursor; | P23847
(DPPA_ECOLI) |
|
|
|
|
|
|
| dsbA | RecName: Full=Thiol:disulfide interchange protein DsbA; Flags: Precursor; | P0AEG4
(DSBA_ECOLI) |
|
|
|
|
|
|
| dut | RecName: Full=Deoxyuridine 5'-triphosphate nucleotidohydrolase; Short=dUTPase; EC=3.6.1.23; AltName: Full=dUTP pyrophosphatase; | P06968
(DUT_ECOLI) |
|
|
|
|
|
|
| eno | RecName: Full=Enolase; EC=4.2.1.11; AltName: Full=2-phospho-D-glycerate hydro-lyase; AltName: Full=2-phosphoglycerate dehydratase; | P0A6P9
(ENO_ECOLI) |
|
|
|
|
|
|
| fabD | RecName: Full=Malonyl CoA-acyl carrier protein transacylase; Short=MCT; EC=2.3.1.39; | P0AAI9
(FABD_ECOLI) |
|
|
|
|
|
|
| fbaA | RecName: Full=Fructose-bisphosphate aldolase class 2; Short=FBP aldolase; Short=FBPA; EC=4.1.2.13; AltName: Full=Fructose-1,6-bisphosphate aldolase; AltName: Full=Fructose-bisphosphate aldolase class II; | P0AB71
(ALF_ECOLI) |
|
|
|
|
|
|
| fbp | RecName: Full=Fructose-1,6-bisphosphatase class 1; Short=FBPase class 1; EC=3.1.3.11; AltName: Full=D-fructose-1,6-bisphosphate 1-phosphohydrolase class 1; | P0A993
(F16PA_ECOLI) |
|
|
|
|
|
|
| fkpA | RecName: Full=FKBP-type peptidyl-prolyl cis-trans isomerase fkpA; Short=PPIase; EC=5.2.1.8; AltName: Full=Rotamase; Flags: Precursor; | P45523
(FKBA_ECOLI) |
|
|
|
|
|
|
| fliY | RecName: Full=Cystine-binding periplasmic protein; Short=CBP; AltName: Full=Protein fliY; AltName: Full=Sulfate starvation-induced protein 7; Short=SSI7; Flags: Precursor; | P0AEM9
(FLIY_ECOLI) |
|
|
|
|
|
|
| fmt | RecName: Full=Methionyl-tRNA formyltransferase; EC=2.1.2.9; | P23882
(FMT_ECOLI) |
|
|
|
|
|
|
| folA | RecName: Full=Dihydrofolate reductase; EC=1.5.1.3; | P0ABQ4
(DYR_ECOLI) |
|
|
|
|
|
|
| frr | RecName: Full=Ribosome-recycling factor; Short=RRF; AltName: Full=Ribosome-releasing factor; | P0A805
(RRF_ECOLI) |
|
|
|
|
|
|
| ftsZ | RecName: Full=Cell division protein ftsZ; | P0A9A6
(FTSZ_ECOLI) |
|
|
|
|
|
|
| fucK | RecName: Full=L-fuculokinase; EC=2.7.1.51; AltName: Full=L-fuculose kinase; | P11553
(FUCK_ECOLI) |
|
|
|
|
|
|
| fur | RecName: Full=Ferric uptake regulation protein; Short=Ferric uptake regulator; | P0A9A9
(FUR_ECOLI) |
|
|
|
|
|
|
| gapA | RecName: Full=Glyceraldehyde-3-phosphate dehydrogenase A; Short=GAPDH-A; EC=1.2.1.12; | P0A9B2
(G3P1_ECOLI) |
|
|
| | {Mi} |
| {Mi,Gm,Aa} |
| {Mi,Gm,Aa} |
|
|
|
| gdhA | RecName: Full=NADP-specific glutamate dehydrogenase; Short=NADP-GDH; EC=1.4.1.4; | P00370
(DHE4_ECOLI) |
|
|
|
|
|
|
| glnA | RecName: Full=Glutamine synthetase; EC=6.3.1.2; AltName: Full=Glutamate--ammonia ligase; | P0A9C5
(GLNA_ECOLI) |
|
|
|
|
|
|
| glnH | RecName: Full=Glutamine-binding periplasmic protein; Short=GlnBP; Flags: Precursor; | P0AEQ3
(GLNH_ECOLI) |
|
|
|
|
|
|
| glnS | RecName: Full=Glutaminyl-tRNA synthetase; EC=6.1.1.18; AltName: Full=Glutamine--tRNA ligase; Short=GlnRS; | P00962
(SYQ_ECOLI) |
|
|
|
|
|
|
| glpD | RecName: Full=Aerobic glycerol-3-phosphate dehydrogenase; EC=1.1.5.3; | P13035
(GLPD_ECOLI) |
|
|
|
|
|
|
| glpK | RecName: Full=Glycerol kinase; EC=2.7.1.30; AltName: Full=ATP:glycerol 3-phosphotransferase; AltName: Full=Glycerokinase; Short=GK; | P0A6F3
(GLPK_ECOLI) |
|
|
|
|
|
|
| glpR | RecName: Full=Glycerol-3-phosphate regulon repressor; | P0ACL0
(GLPR_ECOLI) |
|
|
|
|
|
|
| gltD | RecName: Full=Glutamate synthase [NADPH] small chain; EC=1.4.1.13; AltName: Full=Glutamate synthase subunit beta; Short=GLTS beta chain; AltName: Full=NADPH-GOGAT; | P09832
(GLTD_ECOLI) |
|
|
|
|
|
|
| gltI | RecName: Full=Glutamate/aspartate periplasmic-binding protein; Flags: Precursor; | P37902
(GLTI_ECOLI) |
|
|
|
|
|
|
| glyA | RecName: Full=Serine hydroxymethyltransferase; Short=SHMT; Short=Serine methylase; EC=2.1.2.1; | P0A825
(GLYA_ECOLI) |
|
|
|
|
|
|
| gor | RecName: Full=Glutathione reductase; Short=GR; Short=GRase; EC=1.8.1.7; | P06715
(GSHR_ECOLI) |
|
|
|
|
|
|
| gpmA | RecName: Full=2,3-bisphosphoglycerate-dependent phosphoglycerate mutase; Short=BPG-dependent PGAM; Short=PGAM; Short=Phosphoglyceromutase; Short=dPGM; EC=5.4.2.1; | P62707
(GPMA_ECOLI) |
|
|
|
|
|
|
| grcA | RecName: Full=Autonomous glycyl radical cofactor; | P68066
(GRCA_ECOLI) |
|
|
|
|
|
|
| greA | RecName: Full=Transcription elongation factor greA; AltName: Full=Transcript cleavage factor greA; | P0A6W5
(GREA_ECOLI) |
|
|
|
|
|
|
| groL | RecName: Full=60 kDa chaperonin; AltName: Full=GroEL protein; AltName: Full=Protein Cpn60; | P0A6F5
(CH60_ECOLI) |
|
|
|
|
|
|
| groS | RecName: Full=10 kDa chaperonin; AltName: Full=GroES protein; AltName: Full=Protein Cpn10; | P0A6F9
(CH10_ECOLI) |
|
|
|
|
|
|
| grpE | RecName: Full=Protein grpE; AltName: Full=HSP-70 cofactor; AltName: Full=HSP24; AltName: Full=Heat shock protein B25.3; | P09372
(GRPE_ECOLI) |
|
|
|
|
|
|
| guaB | RecName: Full=Inosine-5'-monophosphate dehydrogenase; Short=IMP dehydrogenase; Short=IMPD; Short=IMPDH; EC=1.1.1.205; | P0ADG7
(IMDH_ECOLI) |
|
|
|
|
|
|
| gyrA | RecName: Full=DNA gyrase subunit A; EC=5.99.1.3; | P0AES4
(GYRA_ECOLI) |
|
|
|
|
|
|
| hdeA | RecName: Full=Chaperone-like protein hdeA; AltName: Full=10K-S protein; Flags: Precursor; | P0AES9
(HDEA_ECOLI) |
|
|
|
|
|
|
| hdeB | RecName: Full=Protein hdeB; AltName: Full=10K-L protein; Flags: Precursor; | P0AET2
(HDEB_ECOLI) |
|
|
|
|
|
|
| hdhA | RecName: Full=7-alpha-hydroxysteroid dehydrogenase; Short=7-alpha-HSDH; EC=1.1.1.159; | P0AET8
(HDHA_ECOLI) |
|
|
|
|
|
|
| hisB | RecName: Full=Histidine biosynthesis bifunctional protein hisB; Includes: RecName: Full=Histidinol-phosphatase; EC=3.1.3.15; Includes: RecName: Full=Imidazoleglycerol-phosphate dehydratase; Short=IGPD; EC=4.2.1.19; | P06987
(HIS7_ECOLI) |
|
|
|
|
|
|
| hisJ | RecName: Full=Histidine-binding periplasmic protein; Short=HBP; Flags: Precursor; | P0AEU0
(HISJ_ECOLI) |
|
|
|
|
|
|
| hisS | RecName: Full=Histidyl-tRNA synthetase; EC=6.1.1.21; AltName: Full=Histidine--tRNA ligase; Short=HisRS; | P60906
(SYH_ECOLI) |
|
|
|
|
|
|
| hldD | RecName: Full=ADP-L-glycero-D-manno-heptose-6-epimerase; EC=5.1.3.20; AltName: Full=ADP-L-glycero-beta-D-manno-heptose-6-epimerase; Short=ADP-glyceromanno-heptose 6-epimerase; Short=ADP-hep 6-epimerase; Short=AGME; | P67910
(HLDD_ECOLI) |
|
|
|
|
|
|
| hns | RecName: Full=DNA-binding protein H-NS; AltName: Full=Histone-like protein HLP-II; AltName: Full=Protein B1; AltName: Full=Protein H1; | P0ACF8
(HNS_ECOLI) |
|
| | 10'782 |
| 15'616 |
| 15'660 |
| 12'496 |
| | {Mi,Aa} |
| {Mi,Gm,Aa} |
| {Gm} |
| {Mi,Gm,Aa} |
|
|
|
| hslU | RecName: Full=ATP-dependent protease ATPase subunit HslU; AltName: Full=Heat shock protein HslU; AltName: Full=Unfoldase HslU; | P0A6H5
(HSLU_ECOLI) |
|
|
|
|
|
|
| hslV | RecName: Full=ATP-dependent protease subunit HslV; EC=3.4.25.2; AltName: Full=Heat shock protein HslV; | P0A7B8
(HSLV_ECOLI) |
|
|
|
|
|
|
| htpG | RecName: Full=Chaperone protein htpG; AltName: Full=Heat shock protein C62.5; AltName: Full=Heat shock protein htpG; AltName: Full=High temperature protein G; | P0A6Z3
(HTPG_ECOLI) |
|
|
|
|
|
|
| hypB | RecName: Full=Hydrogenase isoenzymes nickel incorporation protein hypB; | P0AAN3
(HYPB_ECOLI) |
|
|
|
|
|
|
| hypD | RecName: Full=Hydrogenase isoenzymes formation protein hypD; | P24192
(HYPD_ECOLI) |
|
|
|
|
|
|
| ibpA | RecName: Full=Small heat shock protein ibpA; AltName: Full=16 kDa heat shock protein A; | P0C054
(IBPA_ECOLI) |
|
|
|
|
|
|
| icd | RecName: Full=Isocitrate dehydrogenase [NADP]; Short=IDH; EC=1.1.1.42; AltName: Full=IDP; AltName: Full=NADP(+)-specific ICDH; AltName: Full=Oxalosuccinate decarboxylase; | P08200
(IDH_ECOLI) |
|
|
|
|
|
|
| ilvC | RecName: Full=Ketol-acid reductoisomerase; EC=1.1.1.86; AltName: Full=Acetohydroxy-acid isomeroreductase; AltName: Full=Alpha-keto-beta-hydroxylacil reductoisomerase; | P05793
(ILVC_ECOLI) |
|
|
|
|
|
|
| ilvE | RecName: Full=Branched-chain-amino-acid aminotransferase; Short=BCAT; EC=2.6.1.42; AltName: Full=Transaminase B; | P0AB80
(ILVE_ECOLI) |
|
|
|
|
|
|
| ispA | RecName: Full=Farnesyl diphosphate synthase; Short=FPP synthase; EC=2.5.1.10; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Geranyltranstransferase; | P22939
(ISPA_ECOLI) |
|
|
|
|
|
|
| ivy | RecName: Full=Inhibitor of vertebrate lysozyme; Flags: Precursor; | P0AD59
(IVY_ECOLI) |
|
|
|
|
|
|
| kdsB | RecName: Full=3-deoxy-manno-octulosonate cytidylyltransferase; EC=2.7.7.38; AltName: Full=CMP-2-keto-3-deoxyoctulosonic acid synthase; Short=CKS; Short=CMP-KDO synthase; | P04951
(KDSB_ECOLI) |
|
|
|
|
|
|
| leuC | RecName: Full=3-isopropylmalate dehydratase large subunit; EC=4.2.1.33; AltName: Full=Alpha-IPM isomerase; Short=IPMI; AltName: Full=Isopropylmalate isomerase; | P0A6A6
(LEUC_ECOLI) |
|
|
|
|
|
|
| leuS | RecName: Full=Leucyl-tRNA synthetase; EC=6.1.1.4; AltName: Full=Leucine--tRNA ligase; Short=LeuRS; | P07813
(SYL_ECOLI) |
|
|
|
|
|
|
| livJ | RecName: Full=Leu/Ile/Val-binding protein; Short=LIV-BP; Flags: Precursor; | P0AD96
(LIVJ_ECOLI) |
|
|
|
|
|
|
| livK | RecName: Full=Leucine-specific-binding protein; Short=L-BP; Short=LS-BP; Flags: Precursor; | P04816
(LIVK_ECOLI) |
|
|
|
|
|
|
| lpdA | RecName: Full=Dihydrolipoyl dehydrogenase; EC=1.8.1.4; AltName: Full=Dihydrolipoamide dehydrogenase; AltName: Full=E3 component of pyruvate and 2-oxoglutarate dehydrogenases complexes; AltName: Full=Glycine cleavage system L protein; | P0A9P0
(DLDH_ECOLI) |
|
|
|
|
|
|
| lysA | RecName: Full=Diaminopimelate decarboxylase; Short=DAP decarboxylase; Short=DAPDC; EC=4.1.1.20; | P00861
(DCDA_ECOLI) |
|
|
|
|
|
|
| lysS | RecName: Full=Lysyl-tRNA synthetase; EC=6.1.1.6; AltName: Full=Lysine--tRNA ligase; Short=LysRS; | P0A8N3
(SYK1_ECOLI) |
|
|
|
|
|
|
| maa | RecName: Full=Maltose O-acetyltransferase; EC=2.3.1.79; AltName: Full=Maltose transacetylase; | P77791
(MAA_ECOLI) |
|
|
|
|
|
|
| malE | RecName: Full=Maltose-binding periplasmic protein; AltName: Full=MBP; AltName: Full=MMBP; AltName: Full=Maltodextrin-binding protein; Flags: Precursor; | P0AEX9
(MALE_ECOLI) |
|
|
|
|
|
|
| map | RecName: Full=Methionine aminopeptidase; Short=MAP; EC=3.4.11.18; AltName: Full=Peptidase M; | P0AE18
(AMPM_ECOLI) |
|
|
|
|
|
|
| mdh | RecName: Full=Malate dehydrogenase; EC=1.1.1.37; | P61889
(MDH_ECOLI) |
|
|
| | {Mi,Gm,Aa} |
| {Mi,Aa} |
| {Mi,Gm,Aa} |
|
|
|
| metG | RecName: Full=Methionyl-tRNA synthetase; EC=6.1.1.10; AltName: Full=Methionine--tRNA ligase; Short=MetRS; | P00959
(SYM_ECOLI) |
|
|
|
|
|
|
| metH | RecName: Full=Methionine synthase; EC=2.1.1.13; AltName: Full=5-methyltetrahydrofolate--homocysteine methyltransferase; AltName: Full=Methionine synthase, vitamin-B12-dependent; Short=MS; | P13009
(METH_ECOLI) |
|
|
|
|
|
|
| metK | RecName: Full=S-adenosylmethionine synthase; Short=AdoMet synthase; EC=2.5.1.6; AltName: Full=MAT; AltName: Full=Methionine adenosyltransferase; | P0A817
(METK_ECOLI) |
|
|
|
|
|
|
| mglB | RecName: Full=D-galactose-binding periplasmic protein; Short=GBP; AltName: Full=D-galactose/ D-glucose-binding protein; Short=GGBP; Flags: Precursor; | P0AEE5
(DGAL_ECOLI) |
|
|
|
|
|
|
| mtnN | RecName: Full=5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase; Short=MTA/SAH nucleosidase; Short=MTAN; EC=3.2.2.9; AltName: Full=5'-methylthioadenosine nucleosidase; Short=MTA nucleosidase; AltName: Full=P46; AltName: Full=S-adenosylhomocysteine nucleosidase; Short=AdoHcy nucleosidase; Short=SAH nucleosidase; Short=SRH nucleosidase; | P0AF12
(MTNN_ECOLI) |
|
|
|
|
|
|
| murE | RecName: Full=UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase; EC=6.3.2.13; AltName: Full=Meso-A2pm-adding enzyme; AltName: Full=Meso-diaminopimelate-adding enzyme; AltName: Full=UDP-MurNAc-L-Ala-D-Glu:meso-diaminopimelate ligase; AltName: Full=UDP-MurNAc-tripeptide synthetase; AltName: Full=UDP-N-acetylmuramyl-tripeptide synthetase; | P22188
(MURE_ECOLI) |
|
|
|
|
|
|
| mutS | RecName: Full=DNA mismatch repair protein mutS; | P23909
(MUTS_ECOLI) |
|
|
|
|
|
|
| ndk | RecName: Full=Nucleoside diphosphate kinase; Short=NDK; Short=NDP kinase; EC=2.7.4.6; AltName: Full=Nucleoside-2-P kinase; | P0A763
(NDK_ECOLI) |
|
|
|
|
|
|
| nusA | RecName: Full=Transcription elongation protein nusA; AltName: Full=L factor; AltName: Full=N utilization substance protein A; | P0AFF6
(NUSA_ECOLI) |
|
|
|
|
|
|
| nusG | RecName: Full=Transcription antitermination protein nusG; | P0AFG0
(NUSG_ECOLI) |
|
|
|
|
|
|
| ompA | RecName: Full=Outer membrane protein A; AltName: Full=Outer membrane protein II*; Flags: Precursor; | P0A910
(OMPA_ECOLI) |
|
|
|
|
|
|
| ompF | RecName: Full=Outer membrane protein F; AltName: Full=Outer membrane protein 1A; AltName: Full=Outer membrane protein B; AltName: Full=Outer membrane protein IA; AltName: Full=Porin OmpF; Flags: Precursor; | P02931
(OMPF_ECOLI) |
|
|
|
|
|
|
| oppA | RecName: Full=Periplasmic oligopeptide-binding protein; Flags: Precursor; | P23843
(OPPA_ECOLI) |
|
| | 56'854 |
| 57'529 |
| 56'188 |
| 54'555 |
| | {Mi,Aa} |
| {Mi,Aa} |
| {Mi,Gm,Aa} |
| {Mi,Aa} |
|
|
|
| pfkA | RecName: Full=6-phosphofructokinase isozyme 1; EC=2.7.1.11; AltName: Full=6-phosphofructokinase isozyme I; AltName: Full=Phosphofructokinase 1; AltName: Full=Phosphohexokinase 1; | P0A796
(K6PF1_ECOLI) |
|
|
|
|
|
|
| pfkB | RecName: Full=6-phosphofructokinase isozyme 2; Short=Phosphofructokinase-2; EC=2.7.1.11; | P06999
(K6PF2_ECOLI) |
|
|
|
|
|
|
| pflB | RecName: Full=Formate acetyltransferase 1; EC=2.3.1.54; AltName: Full=Pyruvate formate-lyase 1; | P09373
(PFLB_ECOLI) |
|
|
|
|
|
|
| pgk | RecName: Full=Phosphoglycerate kinase; EC=2.7.2.3; | P0A799
(PGK_ECOLI) |
|
|
|
|
|
|
| pheS | RecName: Full=Phenylalanyl-tRNA synthetase alpha chain; EC=6.1.1.20; AltName: Full=Phenylalanine--tRNA ligase alpha chain; Short=PheRS; | P08312
(SYFA_ECOLI) |
|
|
|
|
|
|
| pheT | RecName: Full=Phenylalanyl-tRNA synthetase beta chain; EC=6.1.1.20; AltName: Full=Phenylalanine--tRNA ligase beta chain; Short=PheRS; | P07395
(SYFB_ECOLI) |
|
|
|
|
|
|
| phoE | RecName: Full=Outer membrane pore protein E; Flags: Precursor; | P02932
(PHOE_ECOLI) |
|
|
|
|
|
|
| pnp | RecName: Full=Polyribonucleotide nucleotidyltransferase; EC=2.7.7.8; AltName: Full=Polynucleotide phosphorylase; Short=PNPase; | P05055
(PNP_ECOLI) |
|
|
|
|
|
|
| polA | RecName: Full=DNA polymerase I; Short=POL I; EC=2.7.7.7; | P00582
(DPO1_ECOLI) |
|
|
|
|
|
|
| potD | RecName: Full=Spermidine/putrescine-binding periplasmic protein; Short=SPBP; Flags: Precursor; | P0AFK9
(POTD_ECOLI) |
|
|
|
|
|
|
| poxB | RecName: Full=Pyruvate dehydrogenase [ubiquinone]; EC=1.2.5.1; AltName: Full=Pyruvate oxidase; Short=POX; Contains: RecName: Full=Alpha-peptide; | P07003
(POXB_ECOLI) |
|
|
|
|
|
|
| ppa | RecName: Full=Inorganic pyrophosphatase; EC=3.6.1.1; AltName: Full=Pyrophosphate phospho-hydrolase; Short=PPase; | P0A7A9
(IPYR_ECOLI) |
|
|
|
|
|
|
| ppiB | RecName: Full=Peptidyl-prolyl cis-trans isomerase B; Short=PPIase B; EC=5.2.1.8; AltName: Full=Rotamase B; | P23869
(PPIB_ECOLI) |
|
|
|
|
|
|
| ppsA | RecName: Full=Phosphoenolpyruvate synthase; Short=PEP synthase; EC=2.7.9.2; AltName: Full=Pyruvate, water dikinase; | P23538
(PPSA_ECOLI) |
|
|
|
|
|
|
| pta | RecName: Full=Phosphate acetyltransferase; EC=2.3.1.8; AltName: Full=Phosphotransacetylase; | P0A9M8
(PTA_ECOLI) |
|
|
|
|
|
|
| ptsH | RecName: Full=Phosphocarrier protein HPr; EC=2.7.11.-; AltName: Full=Histidine-containing protein; | P0AA04
(PTHP_ECOLI) |
|
|
|
|
|
|
| ptsI | RecName: Full=Phosphoenolpyruvate-protein phosphotransferase; EC=2.7.3.9; AltName: Full=Phosphotransferase system, enzyme I; | P08839
(PT1_ECOLI) |
|
|
|
|
|
|
| pyrB | RecName: Full=Aspartate carbamoyltransferase catalytic chain; EC=2.1.3.2; AltName: Full=Aspartate transcarbamylase; Short=ATCase; | P0A786
(PYRB_ECOLI) |
|
|
|
|
|
|
| pyrF | RecName: Full=Orotidine 5'-phosphate decarboxylase; EC=4.1.1.23; AltName: Full=OMP decarboxylase; Short=OMPDCase; Short=OMPdecase; | P08244
(PYRF_ECOLI) |
|
|
|
|
|
|
| pyrI | RecName: Full=Aspartate carbamoyltransferase regulatory chain; | P0A7F3
(PYRI_ECOLI) |
|
|
|
|
|
|
| raiA | RecName: Full=Ribosome-associated inhibitor A; AltName: Full=Protein Y; AltName: Full=SpotY; Short=pY; | P0AD49
(RAIA_ECOLI) |
|
|
|
|
|
|
| rbfA | RecName: Full=Ribosome-binding factor A; AltName: Full=Protein P15B; | P0A7G2
(RBFA_ECOLI) |
|
|
|
|
|
|
| recA | RecName: Full=Protein RecA; AltName: Full=Recombinase A; | P0A7G6
(RECA_ECOLI) |
|
|
|
|
|
|
| rimL | RecName: Full=Ribosomal-protein-serine acetyltransferase; EC=2.3.1.-; AltName: Full=Acetylating enzyme for N-terminal of ribosomal protein L7/L12; | P13857
(RIML_ECOLI) |
|
|
|
|
|
|
| rpiA | RecName: Full=Ribose-5-phosphate isomerase A; EC=5.3.1.6; AltName: Full=Phosphoriboisomerase A; Short=PRI; | P0A7Z0
(RPIA_ECOLI) |
|
|
|
|
|
|
| rplI | RecName: Full=50S ribosomal protein L9; | P0A7R1
(RL9_ECOLI) |
|
|
|
|
|
|
| rplU | RecName: Full=50S ribosomal protein L21; | P0AG48
(RL21_ECOLI) |
|
|
|
|
|
|
| rpoB | RecName: Full=DNA-directed RNA polymerase subunit beta; Short=RNAP subunit beta; EC=2.7.7.6; AltName: Full=RNA polymerase subunit beta; AltName: Full=Transcriptase subunit beta; | P0A8V2
(RPOB_ECOLI) |
|
|
|
|
|
|
| rpsA | RecName: Full=30S ribosomal protein S1; | P0AG67
(RS1_ECOLI) |
|
|
|
|
|
|
| rpsF | RecName: Full=30S ribosomal protein S6; Contains: RecName: Full=30S ribosomal protein S6, fully modified isoform; Contains: RecName: Full=30S ribosomal protein S6, non-modified isoform; | P02358
(RS6_ECOLI) |
|
| | 15'845 |
| 15'865 |
| 15'845 |
| 11'023 |
| | {Mi,Gm,Aa} |
| {Mi,Gm,Aa} |
| {Mi,Gm,Aa} |
| {Mi,Aa} |
| | Expression |
| Expression |
| Expression |
| Expression |
| | 2, 3, 6, 8 |
| 2, 3, 6, 8 |
| 2, 3, 6, 8 |
| 1, 8 |
|
| rsmD | RecName: Full=Ribosomal RNA small subunit methyltransferase D; EC=2.1.1.171; AltName: Full=16S rRNA m2G966 methyltransferase; AltName: Full=rRNA (guanine-N(2)-)-methyltransferase; | P0ADX9
(RSMD_ECOLI) |
|
|
|
|
|
|
| sdhA | RecName: Full=Succinate dehydrogenase flavoprotein subunit; EC=1.3.99.1; | P0AC41
(DHSA_ECOLI) |
|
|
|
|
|
|
| serC | RecName: Full=Phosphoserine aminotransferase; EC=2.6.1.52; AltName: Full=Phosphohydroxythreonine aminotransferase; Short=PSAT; | P23721
(SERC_ECOLI) |
|
|
|
|
|
|
| serS | RecName: Full=Seryl-tRNA synthetase; EC=6.1.1.11; AltName: Full=Serine--tRNA ligase; Short=SerRS; AltName: Full=Seryl-tRNA(Ser/Sec) synthetase; | P0A8L1
(SYS_ECOLI) |
|
|
|
|
|
|
| sodA | RecName: Full=Superoxide dismutase [Mn]; EC=1.15.1.1; AltName: Full=MnSOD; | P00448
(SODM_ECOLI) |
|
|
|
|
|
|
| sodB | RecName: Full=Superoxide dismutase [Fe]; EC=1.15.1.1; | P0AGD3
(SODF_ECOLI) |
|
|
|
|
|
|
| sspA | RecName: Full=Stringent starvation protein A; | P0ACA3
(SSPA_ECOLI) |
|
|
|
|
|
|
| sucA | RecName: Full=2-oxoglutarate dehydrogenase E1 component; EC=1.2.4.2; AltName: Full=Alpha-ketoglutarate dehydrogenase; | P0AFG3
(ODO1_ECOLI) |
|
|
|
|
|
|
| sucB | RecName: Full=Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex; EC=2.3.1.61; AltName: Full=2-oxoglutarate dehydrogenase complex component E2; Short=OGDC-E2; AltName: Full=Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex; | P0AFG6
(ODO2_ECOLI) |
|
|
|
|
|
|
| sucC | RecName: Full=Succinyl-CoA ligase [ADP-forming] subunit beta; EC=6.2.1.5; AltName: Full=Succinyl-CoA synthetase subunit beta; Short=SCS-beta; | P0A836
(SUCC_ECOLI) |
|
|
|
|
|
|
| sucD | RecName: Full=Succinyl-CoA ligase [ADP-forming] subunit alpha; EC=6.2.1.5; AltName: Full=Succinyl-CoA synthetase subunit alpha; Short=SCS-alpha; | P0AGE9
(SUCD_ECOLI) |
|
|
|
|
|
|
| talB | RecName: Full=Transaldolase B; EC=2.2.1.2; | P0A870
(TALB_ECOLI) |
|
|
|
|
|
|
| thrC | RecName: Full=Threonine synthase; Short=TS; EC=4.2.3.1; | P00934
(THRC_ECOLI) |
|
|
|
|
|
|
| tig | RecName: Full=Trigger factor; Short=TF; | P0A850
(TIG_ECOLI) |
|
|
|
|
|
|
| tolB | RecName: Full=Protein tolB; Flags: Precursor; | P0A855
(TOLB_ECOLI) |
|
|
|
|
|
|
| tpiA | RecName: Full=Triosephosphate isomerase; Short=TIM; EC=5.3.1.1; AltName: Full=Triose-phosphate isomerase; | P0A858
(TPIS_ECOLI) |
|
|
|
|
|
|
| tpx | RecName: Full=Thiol peroxidase; EC=1.11.1.-; AltName: Full=Scavengase P20; | P0A862
(TPX_ECOLI) |
|
|
|
|
|
|
| trpA | RecName: Full=Tryptophan synthase alpha chain; EC=4.2.1.20; | P0A877
(TRPA_ECOLI) |
|
|
|
|
|
|
| trpD | RecName: Full=Anthranilate synthase component II; EC=4.1.3.27; Includes: RecName: Full=Glutamine amidotransferase; Includes: RecName: Full=Anthranilate phosphoribosyltransferase; EC=2.4.2.18; | P00904
(TRPG_ECOLI) |
|
|
|
|
|
|
| trxA | RecName: Full=Thioredoxin-1; Short=Trx-1; | P0AA25
(THIO_ECOLI) |
|
|
|
|
|
|
| trxB | RecName: Full=Thioredoxin reductase; Short=TRXR; EC=1.8.1.9; | P0A9P4
(TRXB_ECOLI) |
|
|
|
|
|
|
| tsf | RecName: Full=Elongation factor Ts; Short=EF-Ts; | P0A6P1
(EFTS_ECOLI) |
|
|
|
|
|
|
| tufB | RecName: Full=Elongation factor Tu 2; Short=EF-Tu 2; AltName: Full=P-43; | P0CE48
(EFTU2_ECOLI) |
|
|
|
|
|
|
| tyrS | RecName: Full=Tyrosyl-tRNA synthetase; EC=6.1.1.1; AltName: Full=Tyrosine--tRNA ligase; Short=TyrRS; | P0AGJ9
(SYY_ECOLI) |
|
|
|
|
|
|
| udp | RecName: Full=Uridine phosphorylase; Short=UPase; Short=UrdPase; EC=2.4.2.3; | P12758
(UDP_ECOLI) |
|
|
|
|
|
|
| ulaD | RecName: Full=3-keto-L-gulonate-6-phosphate decarboxylase ulaD; EC=4.1.1.85; AltName: Full=3-dehydro-L-gulonate-6-phosphate decarboxylase; AltName: Full=KGPDC; AltName: Full=L-ascorbate utilization protein D; | P39304
(ULAD_ECOLI) |
|
|
|
|
|
|
| upp | RecName: Full=Uracil phosphoribosyltransferase; EC=2.4.2.9; AltName: Full=UMP pyrophosphorylase; AltName: Full=UPRTase; | P0A8F0
(UPP_ECOLI) |
|
|
|
|
|
|
| uspA | RecName: Full=Universal stress protein A; | P0AED0
(USPA_ECOLI) |
|
|
|
|
|
|
| uspE | RecName: Full=Universal stress protein E; | P0AAC0
(USPE_ECOLI) |
|
|
|
|
|
|
| uspF | RecName: Full=Universal stress protein F; | P37903
(USPF_ECOLI) |
|
|
|
|
|
|
| uspG | RecName: Full=Universal stress protein G; | P39177
(USPG_ECOLI) |
|
|
|
|
|
|
| xthA | RecName: Full=Exodeoxyribonuclease III; Short=EXO III; Short=Exonuclease III; EC=3.1.11.2; AltName: Full=AP endonuclease VI; | P09030
(EX3_ECOLI) |
|
|
|
|
|
|
| yadK | RecName: Full=Uncharacterized protein yadK; | P37016
(YADK_ECOLI) |
|
|
|
|
|
|
| yceB | RecName: Full=Uncharacterized lipoprotein yceB; Flags: Precursor; | P0AB26
(YCEB_ECOLI) |
|
|
|
|
|
|
| ygiN | RecName: Full=Probable quinol monooxygenase ygiN; Short=QuMo; EC=1.-.-.-; | P0ADU2
(YGIN_ECOLI) |
|
|
|
|
|
|
| yjgF | RecName: Full=RutC family protein yjgF; | P0AF93
(YJGF_ECOLI) |
|
|
|
|
|
|
| znuA | RecName: Full=High-affinity zinc uptake system protein znuA; Flags: Precursor; | P39172
(ZNUA_ECOLI) |
|
|
|
|
|
|
| zwf | RecName: Full=Glucose-6-phosphate 1-dehydrogenase; Short=G6PD; EC=1.1.1.49; Contains: RecName: Full=Extracellular death factor; Short=EDF; | P0AC53
(G6PD_ECOLI) |
|
|
|
|
|
|
| | Mapping legends: { Gm } => 'Gel matching', { Mi } => 'Microsequencing', { Aa } => 'Amino acid composition' |
Cited references
| # | PubMed/Medline | Authors | Location |
|---|
| [1] | 9740056 | Tonella L., Walsh B.J., Sanchez J.-C., Ou K., Wilkins M.R., Tyler M., Frutiger S., Gooley A.A., Pescaru I., Appel R.D., Yan J.X., Bairoch A., Hoogland C., Morch F.S., Hughes G.J., Williams K.L., Hochstrasser D.F.; | Electrophoresis 19(1):1960-1971(1998).
|
| [2] | 8740179 | Pasquali C., Frutiger S., Wilkins M.R., Hughes G.J., Appel R.D., Bairoch A., Schaller D., Sanchez J.-C., Hochstrasser D.F.; | Electrophoresis 17(1):547-555(1996).
|
| [3] | - | Vanbogelen R.A., Abshire K.Z., Pertsemlidis A., Clark R.L., Neidhardt F.C.; | (IN) Neidhardt et al. (eds.);
RL Escherichia coli and Salmonella: Cellular and Molecular Biology (2nd ed.), pp.2067-2117, ASM Press, Washington DC (1996).
|
| [4] | 6339477 | Smith M.W., Neidhardt F.C.; | J. Bacteriol. 154(1):344-350(1983).
|
| [5] | 3553157 | Jones P.G., Vanbogelen R.A., Neidhardt F.C.; | J. Bacteriol. 169(1):2092-2095(1987).
|
| [6] | 6988414 | Bloch P.L., Phillips T.A., Neidhardt F.C.; | J. Bacteriol. 141(1):1409-1420(1980).
|
| [7] | 156716 | Herendeen S.L., Vanbogelen R.A., Neidhardt F.C.; | J. Bacteriol. 139(1):185-194(1979).
|
| [8] | 352533 | Pedersen S., Bloch P.L., Reeh S., Neidhardt F.C.; | Cell 14(1):179-190(1978).
|
| [9] | 7002901 | Phillips T.A., Bloch P.L., Neidhardt F.C.; | J. Bacteriol. 144(1):1024-1033(1980).
|
| [10] | 8226654 | Gage D.J., Neidhardt F.C.; | J. Bacteriol. 175(1):7105-7108(1993).
|
| [11] | 3539918 | Vanbogelen R.A., Kelley P.M., Neidhardt F.C.; | J. Bacteriol. 169(1):26-32(1987).
|
| [12] | 6308410 | Neidhardt F.C., Vaughn V., Phillips T.A., Bloch P.L.; | Microbiol. Rev. 47(1):231-284(1983).
|
| [13] | 6339476 | Smith M.W., Neidhardt F.C.; | J. Bacteriol. 154(1):336-343(1983).
|
| [14] | 1346534 | Ernsting B.R., Atkinson M.R., Ninfa A.J., Matthews R.G.; | J. Bacteriol. 174(1):1109-1118(1992).
|
| [15] | 8349554 | Sankar P., Hutton M.E., Vanbogelen R.A., Clark R.L., Neidhardt F.C.; | J. Bacteriol. 175(1):5145-5152(1993).
|
| [16] | 3288628 | Russel M., Model P.; | J. Biol. Chem. 263(1):9015-9019(1988).
|
Gel experimental information
| Information Type | Data | | Map name | ECOLI {Escherichia coli} | | Dimension | 2-D | | Related documents | Sample Preparation / Gel Related Documents
| | Strain Comment [species] | W3110 [Escherichia coli / TaxID:562] | | pI | start: 3.50 - end: 10.00 | | Mw | start: 10000 - end: 200000 | | Number of detected spots | 2364 | | Number of identified spots | 206 | | Number of identified proteins | 180 | | Reference | Bibliographic reference | | Graphical interface |
![[Click for graphical interface]](/swiss-2dpage/data/small-gifs/swiss_2dpage/ECOLI.gif) |
SWISS-2DPAGE (search Map)
|